Team:Glasgow/BiofilmResults
From 2011.igem.org
| Line 15: | Line 15: | ||
</td> | </td> | ||
<td> | <td> | ||
| - | <img src="http://farm7.static.flickr.com/6156/6166741226_e4cfd217bd_m.jpg" /><p><font size="1" color="grey"> | + | <img src="http://farm7.static.flickr.com/6156/6166741226_e4cfd217bd_m.jpg" /><p><font size="1" color="grey">Image 2: 10,000x SEM image of Nissle </br>showing the fimbriae</font></p> |
</td> | </td> | ||
<td> | <td> | ||
| Line 27: | Line 27: | ||
<td> | <td> | ||
<img src="http://farm7.static.flickr.com/6177/6170913398_6e217cc778_m.jpg"/> | <img src="http://farm7.static.flickr.com/6177/6170913398_6e217cc778_m.jpg"/> | ||
| - | <p><font size="1" color="grey">Image | + | <p><font size="1" color="grey">Image 4: 1000x EM of P. aeruginosa biofilm, </br>showing its densely packed structure </br>(courtesy of Dan Walker, University of Glasgow)</font></p> |
</td> | </td> | ||
</tr> | </tr> | ||
| Line 33: | Line 33: | ||
</div> | </div> | ||
</br> | </br> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <img src="http://farm7.static.flickr.com/6156/6171223536_95ab16bc96_z.jpg"> | ||
| + | <p><font size="1" color="grey"> Figure 1: Comparison of RFP E.coli Nissle biofilms to untransformed E.coli Nissle bioflims under light microscope and under excitatory and non-excitatory wavelengths</font></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <td> | ||
| + | <img src="http://farm7.static.flickr.com/6175/6170690387_75d453a871_z.jpg"> | ||
| + | <p><font size="1" color="grey"> Figure 2: Comparison of RFP E.coli Nissle biofilms to puC19 E.coli Nissle biofilms under light microscope and under excitatory and non-excitatory wavelengths</font></p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
<h2>Summary</h2> | <h2>Summary</h2> | ||
Revision as of 02:31, 22 September 2011

Results
The images below show a selection of stages of biofilm formation. Starting with Image 1 showing a lab strain of E.colithat has no fimbriae, and is not forming a biofilm.
Image 2 shows an EM of E.coli Nissle 1917 in the early stages of biofilm formation. The fimbriae that allow the cells to cling to each other are clearly visible.
Image 3 shows a Nissle biofilm in the later stages of formation, with the cells densely packed and the extracellular matrix that holds them together showing.
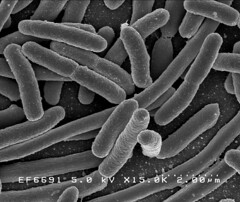
Image 1: 15,000x EM of E.coli for comparison. No fimbriae or EPS is visible. (courtesy of Rocky Mountain Laboratories) |
 Image 2: 10,000x SEM image of Nissle showing the fimbriae |
 Image 3: SEM image of Nissle biofilm showing the extracellular matrix |

Image 4: 1000x EM of P. aeruginosa biofilm, showing its densely packed structure (courtesy of Dan Walker, University of Glasgow) |
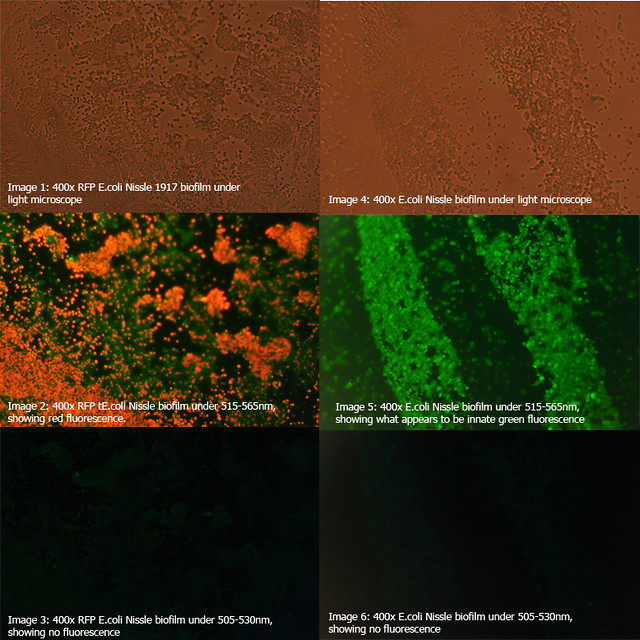
Figure 1: Comparison of RFP E.coli Nissle biofilms to untransformed E.coli Nissle bioflims under light microscope and under excitatory and non-excitatory wavelengths |
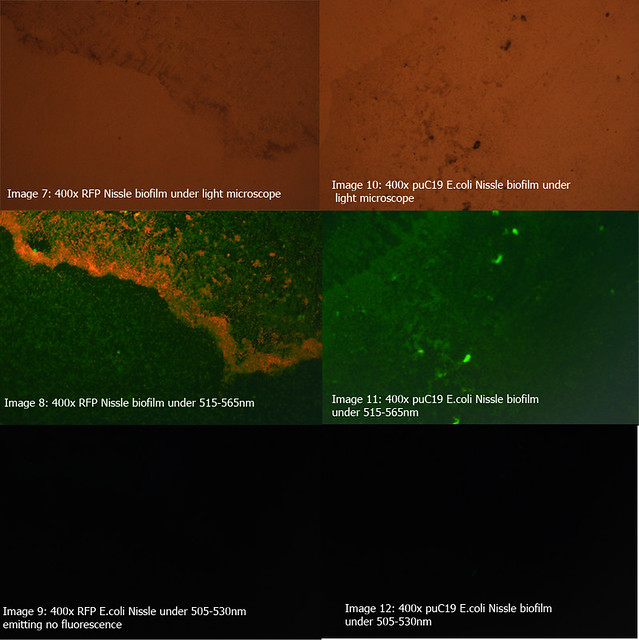
Figure 2: Comparison of RFP E.coli Nissle biofilms to puC19 E.coli Nissle biofilms under light microscope and under excitatory and non-excitatory wavelengths |
Summary
- New Chassis
-Transformable
- Forms biofilms
- Non-pathogenic and compatible with majority of biobricks
-No shuttle vector necessary
-Time series shows that biofilm grows at similar speed to planktonic cells
Well suited for biofilm investigation, especially when intending to transform the biofilm
 "
"
