Team:Wageningen UR/Project/ModelingProj1
From 2011.igem.org
(→Modeling synchronized oscillations) |
(→Modeling synchronized oscillations) |
||
| Line 27: | Line 27: | ||
{{:Team:Wageningen_UR/Templates/NavigationLeft}} | {{:Team:Wageningen_UR/Templates/NavigationLeft}} | ||
{{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | {{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
=== Mathematical model === | === Mathematical model === | ||
| - | |||
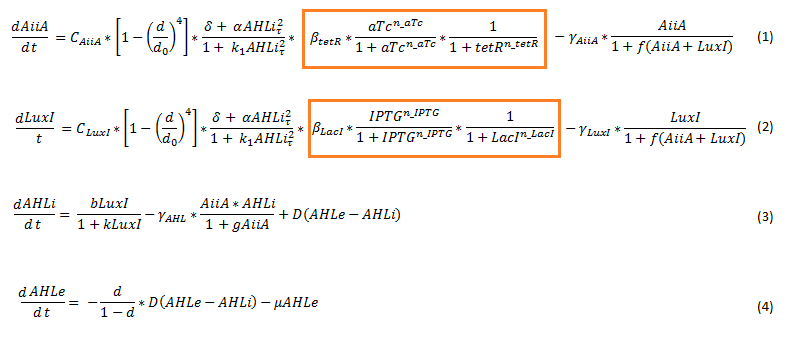
Since our BioBricked oscillatory system is based on the circuit published by Danino et al. in the paper “A synchronized quorum of genetic clocks” [1], our first model of the system is a reproduction of the mathematical model in the supplementary information [2] accompanying the publication mentioned. In their simulations, Danino et al. used a set of four delay differential equations, which we also used as starting point for our modeling work. | Since our BioBricked oscillatory system is based on the circuit published by Danino et al. in the paper “A synchronized quorum of genetic clocks” [1], our first model of the system is a reproduction of the mathematical model in the supplementary information [2] accompanying the publication mentioned. In their simulations, Danino et al. used a set of four delay differential equations, which we also used as starting point for our modeling work. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
The steps (transcription, translation, maturation etc.) from the luxI and aiiA genes to the corresponding proteins are not modeled separately. Instead, the delay of the correlation between the internal AHL concentration, which triggers the expression of the genes, and the corresponding AiiA and LuxI concentrations is simulated by a Hill function. This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex. | The steps (transcription, translation, maturation etc.) from the luxI and aiiA genes to the corresponding proteins are not modeled separately. Instead, the delay of the correlation between the internal AHL concentration, which triggers the expression of the genes, and the corresponding AiiA and LuxI concentrations is simulated by a Hill function. This Hill function takes the history of the system into account, i.e. the concentration of AHL at the time it binds to LuxR to form the activation complex. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
[[Team:Wageningen_UR/Project/ModelingProj1#Modeling_synchronized_oscillations| back to top]] | [[Team:Wageningen_UR/Project/ModelingProj1#Modeling_synchronized_oscillations| back to top]] | ||
| Line 84: | Line 38: | ||
=== Writing a modeling tool in Matlab === | === Writing a modeling tool in Matlab === | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
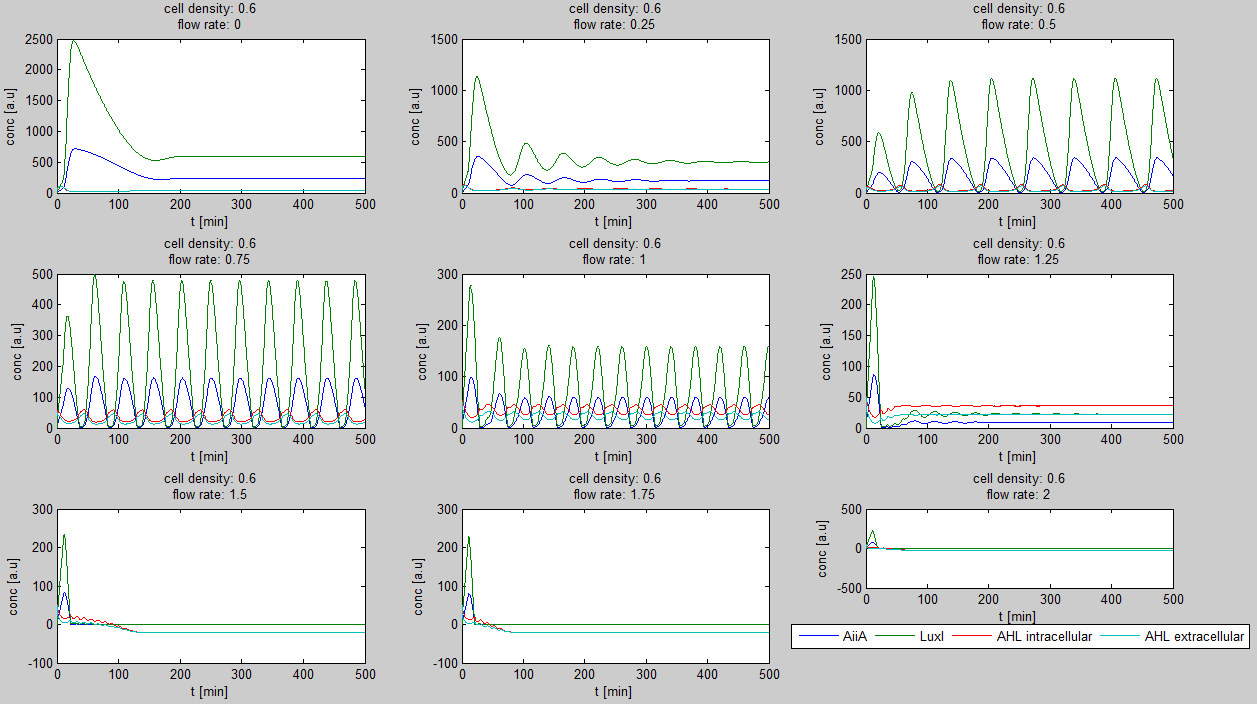
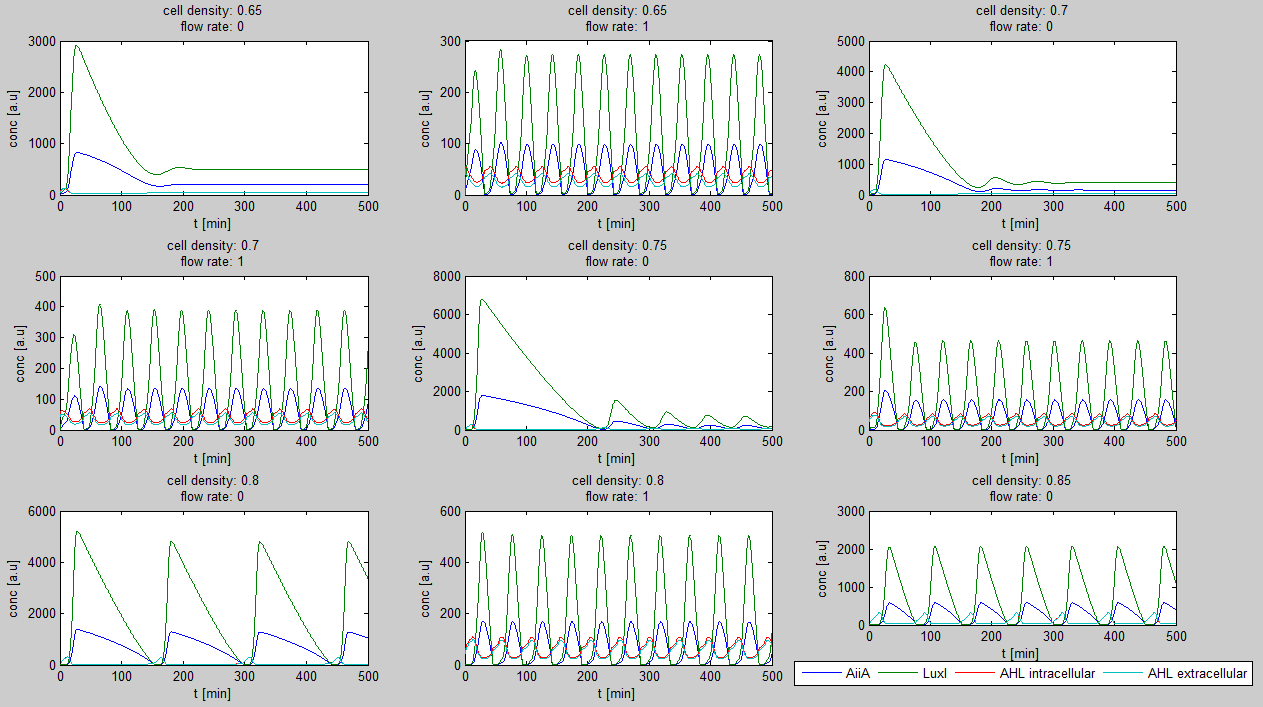
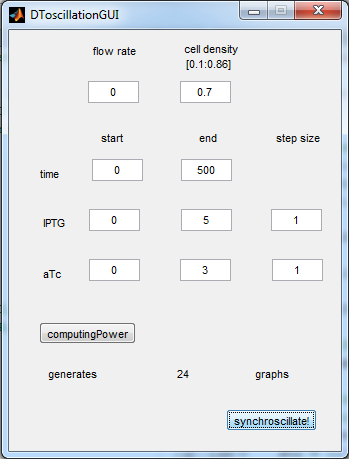
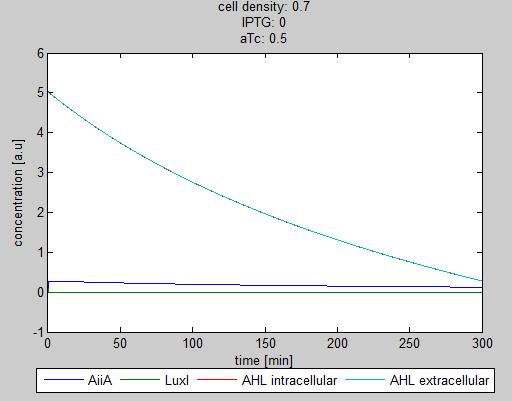
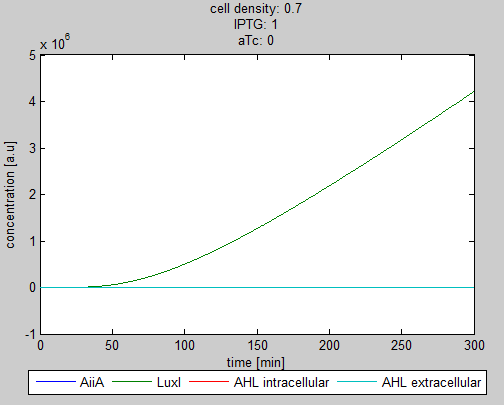
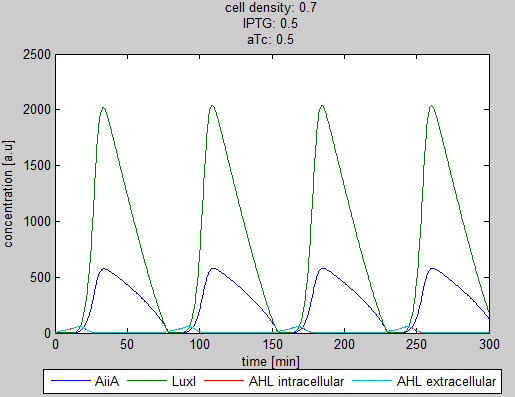
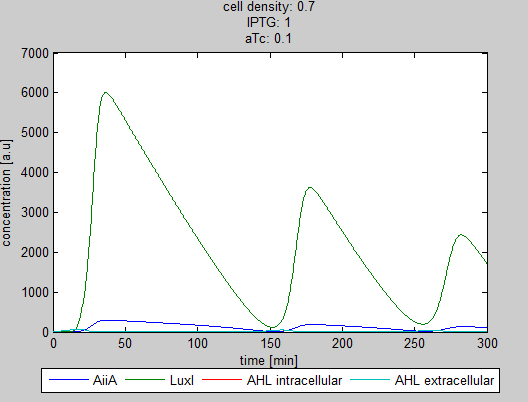
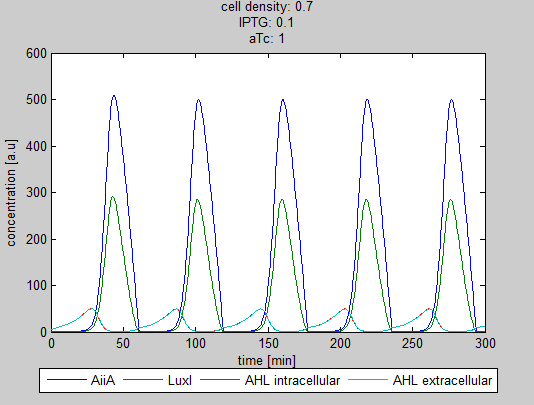
For the first simulations, the same parameters were used as in the cited paper, the only variables thus being the cell density and flow rate. To get an idea about the different conditions in which oscillations could occur, our team created a script for a Matlab modeling tool which uses nested for-loops to vary the flow rate and cell density over a range of values. The resulting tool allows the user to enter the range in which the variables should be varied. The script then iterates over the values and plots graphs of all combinations possible for that range of values. Figure 2 shows an example output of the script. | For the first simulations, the same parameters were used as in the cited paper, the only variables thus being the cell density and flow rate. To get an idea about the different conditions in which oscillations could occur, our team created a script for a Matlab modeling tool which uses nested for-loops to vary the flow rate and cell density over a range of values. The resulting tool allows the user to enter the range in which the variables should be varied. The script then iterates over the values and plots graphs of all combinations possible for that range of values. Figure 2 shows an example output of the script. | ||
| Line 106: | Line 51: | ||
=== Preliminary conclusions for our system === | === Preliminary conclusions for our system === | ||
| - | |||
The first observation from the model was that, for oscillations to occur, the flow rates may not be too fast, especially at lower cell densities (see output graphs above). Since the device used for our system has larger dimensions than the microfluidic device used by Danino et al. the flow rates required could not be achieved by varying height differences alone. <!-- Further information can be found in the information about the [[Team:Wageningen_UR/Project/Devices#Setup| platform used to measure oscillations.]] --> | The first observation from the model was that, for oscillations to occur, the flow rates may not be too fast, especially at lower cell densities (see output graphs above). Since the device used for our system has larger dimensions than the microfluidic device used by Danino et al. the flow rates required could not be achieved by varying height differences alone. <!-- Further information can be found in the information about the [[Team:Wageningen_UR/Project/Devices#Setup| platform used to measure oscillations.]] --> | ||
Revision as of 00:52, 22 September 2011
 "
"