Team:Potsdam Bioware/BioBricks
From 2011.igem.org
UP Sebastian (Talk | contribs) (→BioBrick AraC-TEV) |
(→BioBrick mdnA c-myc gene III) |
||
| Line 310: | Line 310: | ||
<br> | <br> | ||
<b>Full description:</b><br> | <b>Full description:</b><br> | ||
| - | The | + | The following BioBrick is important for testing the suitability of a phage display system as an appropriate screening methoe for recombinant mdnA libraries.<br> |
| - | + | mdnA gene encodes the precursor peptide from the mdn-cluster, which is an essential building block for production of the protease inhibitor microviridin from Microcystis aeruginosa (Ziemert et al., 2008). This gene is known to be able to bind and inhibit proteases. Thus they have a high potential for therapy as they can block disease-relevant proteases (Ziemert et al., 2008 and 2010). <br> | |
| - | The gene III protein is a coat protein from the filamentous bacteriophage M13. It appears only | + | The gene III protein is a coat protein from the filamentous bacteriophage M13. It appears only three to five times on the tip of the phage and is responsible for infection of bacterial cells. In phage display, genes from a DNA library are usually fused to the gene III and are displayed as fusion protein on the surface of the phages. Because it has been shown that the proteins of interest, which are fused to the c-terminus of gene-III can be functionally displayed only the sequence for this part has to be used.<br> |
| - | The inserted | + | The inserted sequence of the myc-tag enables the easy way for detection or purification of the proteins of interest.<br> |
| - | After transformation of E. coli with a vector containing the mdnA-myc-geneIII-fusion gene and co-infection with helper phages E. coli cells were able to produce phage particles | + | After transformation of E. coli with a vector containing the mdnA-myc-geneIII-fusion gene and co-infection with helper phages E. coli cells were able to produce phage particles, which carry the microviridin peptide on their surface. This was confirmed by ELISA test. Using these phages the fundamental suitability of phage display as a screening method for a variety of microviridin variants was proven. |
<br><br> | <br><br> | ||
<b>Source of the part:</b><br> | <b>Source of the part:</b><br> | ||
Revision as of 23:49, 21 September 2011
Contents |
BioBricks
| Label number | BioBrick Nickname | Tube label |
|---|---|---|
| BBa_K627000 | mdnED | 1 |
| BBa_K627001 | mdnA | 2 |
| BBa_K627002 | mdnB | 3 |
| BBa_K627003 | mdnC | 4 |
| BBa_K627004 | mdnD | 5 |
| BBa_K627005 | mdnE | 6 |
| BBa_K627006 | mdnA c-myc gene III | 7 |
| BBa_K627007 | c-myc gene III | 8 |
| BBa_K627008 | AraC TEV protease 1 | 9 |
| BBa_K627009 | AraC TEV protease 2 | 10 |
| BBa_K627010 | AraC TEV protease 3 | 11 |
| BBa_K627011 | AraC 14_3C protease | 12 |
| BBa_K627012 | ssTorA CS-TEV BlaFL | 13 |
| BBa_K627013 | ssTorA CS-14_3C BlaFL | 14 |
| BBa_K627014 | A3_Ara_YFP | 15 |
| BBa_K627015 | A3_lac_YFP | 16 |
BioBrick mdnED
Part name: BBa_K627000
Part type: Coding
Short description: ABC transporter and N-acetyltransferase from the mdn-cluster
Full description:
The BioBrick mdnED is a part of the whole microviridin gene (mdn) cluster, which encodes the protease
inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized
by the cyanobacteria Microcystis aeruginosa (Ziemert et al., 2010). They have a promising potential for therapy as they can block
disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L
biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes,
which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al.,
2008).
In the following BioBrick mdnED the genes mdnD (N-acetyltransferase of the GNAT family) and mdnE (ABC
transporter) is encoded (Ziemert et al., 2008).
Source of the part:
The BioBrick mdnDE as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: TAAATGAATTCGCGGCCGCTTCTAGATGCCTCAATATACTACTAAAC
Reverse primer: ATTTCTGCAGCGGCCGCTACTAGTATCAGCAAACCCTACTTAATTTC
To insert mdnED in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is an expression part, the adenine of mdnE gene's start codon is part of the XbaI
recognition site.
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genebank file:
Media: UP_BioBrick_mdnED.gb
BioBrick mdnA
Part name: BBa_K627001
Part type: Coding
Short description: Ribosomal precursor peptide gene (mdnA) from mdn-cluster
Full description:
The BioBrick mdnA is a part of the whole microviridin gene (mdn) cluster, which encodes the protease inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized by Microcystis aeruginosa (Ziemert et al., 2010). They have a promising potential for therapy as they can block disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes, which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al., 2008).
The following BioBrick mdnA encodes the ribosomal precursor peptide (MdnA), which is essential for microviridin production (Ziemert et al., 2008).
Because this BioBrick is a RFC10 expression part, the adenine of mdnB gene`s start codon is part of the XbaI recognition site.
Source of the part:
The BioBrick mdnA as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: TTCCATGGCGCCAGAGGAATCTAGATGGCATATCCCAACGATC
Reverse primer: CTTCTGACTGGGAAGATTATACCGGTTAATACTAGTAGCGGCCGCTGCAGGACGTC
To insert mdnA in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is an expression part, the adenine of mdnA gene`s start codon is part of the XbaI recognition site. Further the sequence contains a AgeI recognition site after mdnA.
Experiences:
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genbank file:
Media: UP_Biobrick_mdnA.gb
BioBrick mdnB
Part name: BBa_K627002
Part type: Coding
Short description: ATP-grasp-type ligase from mdn-cluster
Full description:
The BioBrick mdnB is a part of the whole microviridin gene (mdn) cluster, which encodes the protease inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized by Microcystis aeruginosa (Ziemert et al., 2010). They have a promising potential for therapy as they can block disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes, which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al., 2008).
The following BioBrick mdnB encodes a ATP-grasp-type ligase (Ziemert et al., 2008).
Because this BioBrick is an expression part, the adenine of mdnB gene`s start codon is part of the XbaI recognition site.
Source of the part:
The BioBrick mdnB as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: ATTATGAATTCGCGGCCGCTTCTAGATGAAAGAATCGCCTAAAGTTG
Reverse primer: TAATCTGCAGCGGCCGCTACTAGTATCAACCGAAGACTAAAAAATCAGCG
To insert mdnB in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is an expression part, the adenine of mdnE gene's start codon is part of the XbaI recognition site.
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genbank file:
Media: UP_BioBrick_mdnB.gb
BioBrick mdnC
Part name: BBa_K627003
Part type: Coding
Short description: ATP-grasp-type ligase from the mdn-cluster
Full description:
The BioBrick mdnC is a part of the whole microviridin gene (mdn) cluster, which encodes the protease
inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized
by Microcystis (Ziemert et al., 2010). They have a promising potential for therapy as they can block
disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L
biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes,
which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al.,
2008).
The following BioBrick mdnC encodes the ATP-grasp-type ligase (Ziemert et al., 2008).
Because this BioBrick is a RF10 expression part, the adenine of mdnC gene`s start codon is part of the XbaI
recognition site.
Source of the part:
The BioBrick mdnC as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: TATTTGAATTCGCGGCCGCTTCTAGATGACCGTTTTAATTGTTAC
Reverse primer: ATTTCTGCAGCGGCCGCTACTAGTATTATGAGTTAACTAGGATTTC
To insert mdnC in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is a RF10 expression part, the adenine of mdnA gene`s start codon is part of the XbaI recognition site.
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genebank file:
Media: UP_BioBrick_mdnC.gb
BioBrick mdnD
Part name: BBa_K627004
Part type: Coding
Short description: N-acetyltransferase from the mdn-cluster
Full description:
The BioBrick mdnD is a part of the whole microviridin gene (mdn) cluster, which encodes the protease
inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized
by Microcystis (Ziemert et al., 2010). They have a promising potential for therapy as they can block
disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L
biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes,
which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al.,
2008).
The following BioBrick mdnD encodes a N-acetyltransferase of the GNAT family (Ziemert et al., 2008).
Because this BioBrick is a RF10 expression part the adenine of mdnD gene's start codon is part of the XbaI
recognition site.
Source of the part:
The BioBrick mdnD as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: TATATGAATTCGCGGCCGCTTCTAGATGAAAGCACTGGAAAAACTG
Reverse primer: ATTTCTGCAGCGGCCGCTACTAGTATCAGCAAACCCTACTTAATTTC
To insert mdnD in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is a RF10 expression part the adenine of mdnD gene`s start codon is part of the XbaI recognition site.
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genebank file
Media: UP_BioBrick_mdnD.gb
BioBrick mdnE
Part name: BBa_K627005
Part type: Coding
Short description: ABC transporter from the mdn-cluster
Full description:
The BioBrick mdnE is a part of the whole microviridin gene (mdn) cluster, which encodes the protease
inhibitor microviridin L. Microviridins are tricyclic depsipeptides, which are ribosomally synthesized
by Microcystis (Ziemert et al., 2010). They have a promising potential for therapy as they can block
disease-relevant proteases (Ziemert et al., 2008).
Microviridins are synthesized from a ribosomal precursor peptide (MdnA). Additionally, the microviridin L
biosynthesis gene cluster consists of genes encoding an ATP-grasp-type ligase (mdnB and mdnC) and genes,
which encode an ABC transporter (mdnE) and a N-acetyltransferase of the GNAT family (mdnD) (Ziemert et al.,
2008).
The following BioBrick mdnE encodes an ABC transporter (Ziemert et al., 2008).
Because this BioBrick is a RF10 expression part the adenine of mdnE gene`s start codon is part of the XbaI
recognition site.
Source of the part:
The BioBrick mdnE as a part of the microviridin gene (mdn) cluster was isolated from Microcystis aeruginosa strain NIES-843.
Design information:
This BioBrick was built by PCR using the following PCR primers
Forward primer: TAAATGAATTCGCGGCCGCTTCTAGATGCCTCAATATACTACTAAAC
Reverse primer: ATTTCTGCAGCGGCCGCTACTAGTACTATATTCTCACCCATTTTAAG
To insert mdnE in the vector pSB1C3, the resulting PCR product and the vector were digested with the restriction enzymes EcoRI and SpeI.
Because this BioBrick is a RF10 expression part the adenine of mdnE gene`s start codon is part of the XbaI recognition site.
References:
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genebank file
Media: UP_BioBrick_mdnE.gb
BioBrick mdnA c-myc gene III
Part name: BBa_K627006
Part type: Coding
Short description: Fusion part of mdnA gene (from mdn-cluster) with c-terminal myc-tag and gene III
Full description:
The following BioBrick is important for testing the suitability of a phage display system as an appropriate screening methoe for recombinant mdnA libraries.
mdnA gene encodes the precursor peptide from the mdn-cluster, which is an essential building block for production of the protease inhibitor microviridin from Microcystis aeruginosa (Ziemert et al., 2008). This gene is known to be able to bind and inhibit proteases. Thus they have a high potential for therapy as they can block disease-relevant proteases (Ziemert et al., 2008 and 2010).
The gene III protein is a coat protein from the filamentous bacteriophage M13. It appears only three to five times on the tip of the phage and is responsible for infection of bacterial cells. In phage display, genes from a DNA library are usually fused to the gene III and are displayed as fusion protein on the surface of the phages. Because it has been shown that the proteins of interest, which are fused to the c-terminus of gene-III can be functionally displayed only the sequence for this part has to be used.
The inserted sequence of the myc-tag enables the easy way for detection or purification of the proteins of interest.
After transformation of E. coli with a vector containing the mdnA-myc-geneIII-fusion gene and co-infection with helper phages E. coli cells were able to produce phage particles, which carry the microviridin peptide on their surface. This was confirmed by ELISA test. Using these phages the fundamental suitability of phage display as a screening method for a variety of microviridin variants was proven.
Source of the part:
The BioBrick mdnA is a part of the microviridin gene (mdn) cluster which was isolated from Microcystis aeruginosa strain NIES-843.
The gene III protein is a coat protein from the filamentous bacteriophage M13.
Design information:
Gene III was amplified from the vector pak100blaKDIR using the following primers
Forward:TAAGCTTCTAGATGGCCGGCGAGCAGAAGCTGATCTCTGAGGAAGACCTGGGTGGTGGCTCTGGTTCC
Reverse: TGCTTAGACGTCCTGCAGCGGCCGCTACTAGTATTAACCGGTAGACTCCTTATTACGCAGTA
The mdnA gene was amplified from the vector pARW089 using the following primers
Forward: TTCCATGGCGCCAGAGGAATCTAGATGGCATATCCCAACGATC
Reverse: CTTCTGACTGGGAAGATTATACCGGTTAATACTAGTAGCGGCCGCTGCAGGACGTC
Gene III was amplified from pak100blaKDIR and mdnA from pARW089 by PCR. The primers were designed to enable the introduction of iGEM. The PCR product gene III was digested by whereas the PCR product mdnA was digested by AgeI. Thus a mdnA-gene III fusion part according to RFC25 was generated whereby AgeI and NgoMIV overhangs are compatible and placed in frame with the protein sequence. The ligation of AgeI and NgoMIV overhangs resulted in a scar coding for the threonine and glycine. Because the introduction of restriction sites before mdnA leaded to a great distance between ribosome binding site and mdnA a second RBS was inserted among SfoI and XbaI recognition sites to ensure a sufficiently expression rate of the mdnA-gene III-fusion gene. Between mdnA and gene III myc sequence was inserted.
Because this BioBrick is an expression part, the adenine of mdnA gene's start codon is part of the XbaI recognition site. Further the sequence contains a AgeI recognition site after gene III.
Experiences:
Detection of phages carrying mdnA on their surface by ELISA
The successful expression of the mdnA-myc-gene III-fusion protein on the surface of the phage was detected by ELISA. Therefore E. coli cells strains XL1-Blue and ER2738 were first transformed with the phagemid pPDV089 before they were infected with helper phages. E. coli cells containing both plasmids were selected. An ELISA test was performed to determine whether these cells are able to produce phage particles carrying the mdnA peptide on their surface. To perform this test anti-c-myc-antibodies were immobilized on ELISA plates and incubated with purified phages. For detection a second antibody coupled with horse radish peroxidase (HRP) was used which binds the gene VIII protein of the phages. The HRP substrate 3,3'-5,5'-tetramethylbenzidine (TMB) was added and in case of binding a colour reaction was expected. The colour shift from achromatic to yellow in wells incubated with phages produced in XL1-blue cells showed the successful expression of mdnA-c-myc-gene III-fusion protein on the phages.In wells incubated with infected ER2738 cells no colour change was obserevd. That might be due to the fact that a mistake during phage purification was noticed.
For more precise results the absorption from 450 – 600 nm was measured. The data were presented in a bar plot. As a negative control helper phages were added instead of produced phages. Furthermore two wells were prepared were the secondary antibody was not added.
The graphic shows clearly the much higher absorption measured in wells, which were incubated with phage particles of interest produced in XL1-Blue cells. As has already pointed out this shows the succeeded expression of mdnA-c-myc-gene III-fusion protein on the surface of the phages.
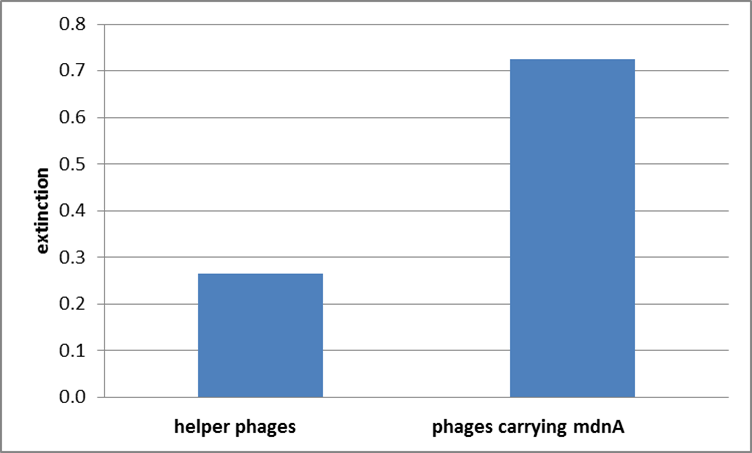
Testing phage display with unmodified mdnA to examine its suitability as screening method
To test the fundamental suitability of this screening method, phages representing unmodified mdnA on their surface and phages not representing mdnA (helper phages) in a ratio of one to one were incubated with immobilized trypsin which is known as a target of mdnA. The display was conducted in ELISA plates. The bound phages were eluted using a buffer with low pH value and neutralized afterwards. To check how many phages interacted with trypsin, E. coli cells XL1-Blue were re-infected with eluted phages and plated on agar with different antibiotics. Cells infected with phages carrying mdnA are able to grow on agar with ampicillin whereas cells infected with helper phages are able to grow on agar with kanamycin. To control the success of the panning round additionally E. coli cells were infected with phage mix before panning and plated on agar. Subsequent the number of clones grew on ampicillin and kanamycin before and after panning was compared. During the running of this step it was noticed that much more cells were infected with helper phages than with phages carrying mdnA despite of the engaged 1:1 ratio. This was surprising and indicated that mdnA on the surface of the phages may inhibit their infectivity. After controlling the plates an infection ratio of phages carrying mdnA to helper phages of 1:400 was calculated. This fact should be analyzed in further experiments.
The results of the first phage display are plotted in the figure below. After one panning round an enrichment of phages carrying mdnA was expected. This is attributable to the fact that phage particles carrying mdnA c-myc geneIII fusion protein on their surface are expected to bind specifically to the immobilized trypsin. Unfortunately this was not observed in this experiment. The ratio of cells growing on kanamycin agar before (4000) to cells growing on kanamycin agar (cells containing helper phages) after panning (750) was determined as 5:1. The ratio of cells growing on ampicillin agar before (12) to cells growing on ampicillin agar (cells containing mdnA carrying phages) after panning (2) was nearly equal. Thus no enrichment of mdnA carrying phages occurred in the first experiment. So it was decided to repeat this experiment under improved conditions. Therefor the number of washing steps during the described experimental procedure was increased. Here the ratio of cells growing on kanamycin agar before (3000) to cells growing on kanamycin agar (cells containing helper phages) after panning (29) was determined as 103:1. The ratio of cells growing on ampicillin agar before (26) to cells growing on ampicillin agar (cells containing mdnA carrying phages) after panning (2) was determined as 13:1. Thus an enrichment factor of eight was reached for the phages displaying mdnA on their surface.
These results indicate that the unmodified mdnA expressed on the phages binds specifically to the immobilized trypsin. Therefore it can be deduced that mdnA is presented in a functional 3D structure. These findings suggest that phage display in general is an appropriate method for screening a recombinant mdnA library. Further experiments are required to optimize this system.
References:
Fuh G., Sidhu S.S. (2000). Efficient phage display of polypeptides fused to the carboxy-terminus of the M13 gene-3 minor coat protein. FEBS Lett. 480(2-3):231-4
Rakonjac J., Feng J., Model P. (1999). Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J Mol Biol. 289(5):1253-65 Smith, G.P. (1985). Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virus surface. Science 228: 1315-17
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
Genebank file:
Media: UP_Biobrick_mdnA_c-myc_geneIII.gb
Gene III
BioBrick AraC-TEV
Part name: BBa_K627008, BBa_K627009, BBa_K627010
Part type: Device
Short description: Fusion part of pBAD arabinose-inducible induction system and the TEV protease
Full description:
Introduction
This BioBrick is a 2 part fusion of arabinose-inducible induction system and the TEV protease.
General Information of the single sub parts:
TEV protease
TEV protease is the common name for the 27 kDa catalytic domain of the Nuclear Inclusion a endopeptidase (NIa) encoded by the tobacco etch virus (TEV). It recognizes a linear epitope of the general form E-Xaa-Xaa-Y -Xaa-Q-(G/S), with cleavage occurring between Q and G or Q and S. In TEV protease the serine nucleophile of the conventional Ser-Asp-His triad is a cysteine instead. This probably explains why TEV protease is resistant to many commonly used protease inhibitors.
At 37°C, the TEV protease forms inclusion body, which leads to an inactive form. Incubated at 30 °C, the protease is expressed as soluble type and is highly active.
Arabinose-inducible induction system
The arabinose-inducible induction system was amplified via PCR from the pBAD_iGEMexpress vector. Based on the inhibitionary funtion of the catabolic activator protein (CAP) it got a very low expression rate without induction by arabinose. For the induction concentration between 2 mM up to 50 mM arabinose can be used to get an high expression rate.
General Function:
When arabinose is added to the media (2mM up to 50 mM), the CAP lost its inhibitory function and the arabinose-inducible induction system gets activated. The protein, here the TEV protease, after the T7 RBS gets express at a very high rate. So we can assume that this system is an effectiv protease generator.
Performance and Summary:
In combination with our created ssTorA_CS-TEV_blaFL device (BBa_K627012, [http://partsregistry.org/Part:BBa_K627012 More Information]) this protease generator was able to mediate the cell death of our transformed E.coli cells at ampicillin concentration up to 100 µg/ml.
Source of the part:
Arabinose inducible operon form pBAD_iGEM_express, TEV protease from Gunther Stier et al.
Design Notes
This BioBrick was built by PCR using the following PCR primers:
Primer used for site directed mutagenesis:
Fragment 1:
- r_TEV_ACCAGC: GGAATGTGCAGCTGGTGTCTGACACC
- f_TEV_iGEM: ATATAGAATTCGCGGCCGCTTCTAGATGGGAGAAAGCTTGTTTAAGGGA
Fragment 2:
- r_TEV_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTATTGCGAGTACACCAATTCATTCAT
- f_TEV_ACCAGC: GGTGTCAGACACCAGCTGCACATTCC
Primer used for assembly PCR of mutated fragments:
- f_TEV_iGEM: ATATAGAATTCGCGGCCGCTTCTAGATGGGAGAAAGCTTGTTTAAGGGA
- r_TEV_iGEM_BamHI: TATAGGATCCACTGCAGCGGCCGCTACTAGTTTATTGCGAGTACACCAATTCATTCAT
Primers used for amplification of pBAD arabinose-inducible induction system:
- f_AraC_iGEM_HindIII: TATAAGCTTGAATTCGCGGCCGCTTCTAGATTATGACAACTTGACGGCTACATCATT
- r_AraC_NgoMIV: ATAGCCGGCCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGCCC
Primers used for amplification and modification of TEV protease:
- f_TEV_AraFusion_NgoMIV: ATATTGCCGGCATGGGAGAAAGCCTGTTTAAGGGA
- r_TEV_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTATTGCGAGTACACCAATTCATTCAT
Experience:
References:
Cabrita, L. D., Gilis, D., Robertson, A. L., Dehouck, Y., Rooman, M. and Bottomley, S. P. (2007). Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 16: 2360-2367
Kapust, R. B., Tözsér, J., Fox, J. D., Anderson, D. E., Cherry, S., Copeland, T. D., and Waugh, D. S. (2001). Tobacco etch virus protease: Mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Prot. Eng. 14: 993-1000.
Lucast, L. J., Batey, R. T., and Doudna, J. A. (2001). Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30: 544-550.
Genebank file
Media:BioBrick_AraC-TEV_comp.gb
Three parts are existing.
BioBrick AraC-14_3C protease
Part name: BBa_K627011
Part type:Device
Short description: Fusion part of arabinose-inducible induction system and the HRV 14_3C protease
Full description:
This BioBrick is a 2 part fusion of arabinose-inducible induction system and the HRV 14_3C protease. The recombinant type 14_3C protease from human rhinovirus (HRV 3C) recognizes the same cleavage site as the native enzyme: LeuGluValLeuPheGln↓GlyPro.
The small, 22 kDa size of the protease got its optimal activity at 4°C but is still very active at 37°C. It is commonly used for an easy tag removal after the purification of recombinant proteins carrying his-tag. The 14_3C works with a catalytic triade, containing the amino acid residues Ser-Asp-His at its active site.
The arabinose-inducible induction system was amplified via PCR from the pBAD_iGEMexpress vector and fused via NgoMIV with the HRV 14_3C protease. An effective induction can be accomplished with 2 mM up to 25 mM arabinose.
Source of the part:
Arabinose inducible operon form pBAD_iGEM_express was ampilied via PCR, 14_3C protease was isolated from the genome of human rhinovirus 14_3C and mutated with primers listed in part description BBa: to remove all iGEM restriction sites inside the protease gene.
Design Notes:
This BioBrick was built by PCR using the following PCR primers:
Site directed mutagenesis of HRV 14 3C protease:
Fragment 1:
- f_14_3C_iGEM: ATATAGAATTCGCGGCCGCTTCTAGATGGGACCAAACACAGAATTTGCACTATCC
- r_14_3C_ACCAGC: ACCATTTGCTGGTACATCATCACCAGG
Fragment 2:
- f_14_3C_ACCAGC: CCTGGTGATGATGTACCAGCAAATGGT
- r_14_3C_tm_XbaI208_A-T: CACTGTAAGCTCAAGATTAATGTTCTC
Fragment 3:
- f_14_3C_tm_Xba208_A-T: GAGAACATTAATCTTGAGCTTACAGTG
- r_14_3C_tm_XbaI280_A-T: ATCCACACCTTCGAGATCTTCTGATAT
Fragment 4:
- f_14_3C_tm_Xba280_A-T: ATATCAGAAGATCTCGAAGGTGTGGAT
- r_14_3C_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTTGTTTCTCTACAAAATATTGTTTTTTAAGTTGAGCTGA
Primers used for first assembly PCR of mutated fragments:
Fragment 5, containing Fragment 1 and 2:
- f_14_3C_iGEM: ATATAGAATTCGCGGCCGCTTCTAGATGGGACCAAACACAGAATTTGCACTATCC
- r_14_3C_tm_Xba208_A-T: CACTGTAAGCTCAAGATTAATGTTCTC
Fragment 6, containing Fragment 3 and 4:
- f_14_3C_tm_Xba208_A-T: GAGAACATTAATCTTGAGCTTACAGTG
- r_14_3C_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTTGTTTCTCTACAAAATATTGTTTTTTAAGTTGAGCTGA
Primer used for final assembly PCR of mutated fragments:
Complete mutated 14_3C protease, containing Fragment 5 and 6:
- f_14_3C_iGEM: ATATAGAATTCGCGGCCGCTTCTAGATGGGACCAAACACAGAATTTGCACTATCC
- r_14_3C_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTTGTTTCTCTACAAAATATTGTTTTTTAAGTTGAGCTGA
Primers used for amplification of the pBAD arabinose-inducible induction system:
- f_AraC_iGEM_HindIII: TATAAGCTTGAATTCGCGGCCGCTTCTAGATTATGACAACTTGACGGCTACATCATT
- r_AraC_NgoMIV: ATAGCCGGCCTCCTTCTTAAAGTTAAACAAAATTATTTCTAGCCC
Primers used for amplification and modification of HRV 14 3C protease:
- f_14_3C_AraFusion_NgoMIV: ATATTGCCGGCATGGGACCAAACACAGAATTTGCACTATCC
- r_14_3C_iGEM_BamHI: ATATAGGATCCACTGCAGCGGCCGCTACTAGTTTTGTTTCTCTACAAAATATTGTTTTTTAAGTTGAGCTGA
Experience:
References:
Combination of Two Separate Binding Domains Defines Stoichiometry between Type III Secretion System Chaperone IpgC and Translocator Protein IpaB, Ravi Kumar Lokareddy, Michele Lunelli, Björn Eilers, Vivien Wolter, and Michael Kolbe, December 17, 2010 The Journal of Biological Chemistry, 285, 39965-39975.
Cleavage of Small Peptides In Vitro by Human Rhinovirus 14 3C Protease expressed in Escherichia coli, MICHAEL G. CORDINGLEY,* R. BRUCE REGISTER, PIA L. CALLAHAN, VICTOR M. GARSKY,AND RICHARD J. COLONNO JOURNAL OF VIROLOGY, Dec. 1989, p. 5037-5045
Genebank file:
Media:BioBrick_AraC-14_3C.gb
BioBrick ssTorA_CS-TEV_blaFL
Part name: BBa_K627012
Part type: Composite Part
Short description: Fusion of ssTorA, a cleavage site for TEV protease and beta lactamase
Full description:
This construct contains the cleavage site for the TEV protease flanked by the sequence of beta-lactamase and the TorA signal sequence for the TAT (twin arginine translocation) pathway, thus in case of the expression of the complete construct the beta lactamase can be transported into periplasm and provide ampicillin resistance. The TorA signal-sequence originates from the enzyme Trimethylamin-N-Oxid-Reduktase, which needs to be folded in the cytoplasm. The beta lactamase is an enzyme which cleaves lactam rings and makes bacteria resistant to antibiotics like ampicillin and penicillin. The cleavage site for the TEV protease was created via oligo hybridization and ligated into the construct with cleavage site for XhoI and NheI, which makes this part modular and easy adaptable for any other protease.
Source
TorA signal sequence and the lactamase was amplified via PCR from the vector pJC354, which originates from two iGEM compatible BioBricks: BBa_K208005 and BBa_I757010. The cleavage site was created, based on the information of the Brenda enzymes database, via 2 oligonucleotides ordered from SigmaAldrich.
Design Notes:
This BioBrick was built by PCR using the following PCR primers:
- p_TorA-f: CTTCTAGATGAACAATAACGATCTCTTTCAGGCATC
- o_Bla_igem_r: CTACTAGTATTAACCGGTCCAATGCTTAATCAGTGAGGCAC
Experience
References
ssTorA - http://partsregistry.org/Part:BBa_K208005
blaFL - http://partsregistry.org/Part:BBa_I757010
Genebank file
Media:BioBrick_ssTorA_CS-TEV_blaFL.gb
BioBrick ssTorA_CS-14_3C_blaFL
Part name: BBa_K627013
Part type: Composite Part
Short description: Fusion of ssTorA, a cleavage site for HRV 14_3C protease and beta lactamase
Full description:
This construct contains the cleavage site for the HRV 14_3C protease flanked by the sequence of beta-lactamase and the TorA signal sequence for the TAT (twin arginine translocation) pathway, thus in case of the expression of the complete construct the beta lactamase can be transported into periplasm and provide ampicillin resistance. The TorA signal-sequence originate from the enzyme Trimethylamin-N-Oxid-Reduktase, which needs to be folded in the cytoplasm. The beta lactamase is an enzyme which cleaves lactam rings and makes bacteria resistant to antibiotics like ampicillin and penicillin. The cleavage site for the HRV 14_3C protease was created via oligo hybridization and ligated into the construct with cleavage site for XhoI and NheI, which makes this part modular and easy adaptable for any other protease.
Source
TorA signal sequence and the lactamase was amplified via PCR from the vector pJC354, which originates from two iGEM compatible BioBricks: BBa_K208005 and BBa_I757010. The cleavage site was created, based on the information of the Brenda enzymes database, via 2 oligonucleotides ordered from SigmaAldrich.
Design Notes:
This BioBrick was built by PCR using the following PCR primers:
- p_TorA-f: CTTCTAGATGAACAATAACGATCTCTTTCAGGCATC
- o_Bla_igem_r: CTACTAGTATTAACCGGTCCAATGCTTAATCAGTGAGGCAC
Experience
References
ssTorA - http://partsregistry.org/Part:BBa_K208005
blaFL - http://partsregistry.org/Part:BBa_I757010
Genebank files
Media:BioBrick_ssTorA_CS-14_3C_blaFL.gb
 "
"
