green light receptor
Digestion for 2A-assembly
Investigators: Julia, Sandra
Digestion of:
|
| vector a
| vector b
| insert 1
| insert 2
| insert 3
| insert 4
|
|
| S2b
| S4
| S64 (Ccas)
| S46 (ho1)
| S50 (pcyA)
| S67 (Ccar)
|
| Enzyme 1
| SpeI
| SpeI
| XbaI
| XbaI
| XbaI
| XbaI
|
| Enzyme 2
| PstI
| PstI
| PstI
| PstI
| PstI
| PstI
|
total volume was 50 µl , 1 µl Alcalic Phosphotase and its buffer
was added after 3 hours of incubation to the vector-plasmids S2 and S4
Ligation
Ligation of the parts:
in vector S2b, which is part BBa_J23110(strong promotor):
in vector S4,which is part BBa_B0034(RBS):
It was incubated over night at 16°C. In the morning ligase was inactivated at 80°C.
Transformation on 1.09.2011
for results see 3.09.2011
blue light receptor
Miniprep
Investigators: Sandra
Miniprep of:
- ♥-A3 Not 1
- ♥-A3 noT 2
- ♥-A3 noT 3
Plates with tetracyclin showed not a single colony.
Testdigest
Investigators: Sandra
Testdigest of minipreps.
Digested with EcoRI and PstI.
Testdigestd showed no bands for the inserts.
new Ligation
Investigators: Sandra
New Ligation with the already digested parts. New digested vector from Julia, pSB1C3.
Parts were ligated for 2 hours this time.
Parts:
- Notgate
- Lovtap
- vector: pSB1C3
Trafo
Investigators: Sandra
After the ligation a transformation was performed and the plates were incubated over night at 37°C. Hopefully we get something to see tomorrow:-)
Plan B: We designed new primers for the gibson assembly.
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
PCR
| Name:
Rüdiger
| Date: 31.08.
|
| Continue from Experiment Gibson Assembly (Date) 30.08.
(Name) Rüdiger
|
| Project Name: Precipitator
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
|
|
| 2.5µl
| Primer dw
|
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
|
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
Gibson did not work because DNA concentration was too high.
| Name
Rüdiger
| Date:
31.08.2011
|
| Continue from Experiment Gibson-Assembly (Date) 30.08. (repeated)
(Name) Rüdiger
|
| Project Name: Blue light receptor: Ligation of Lovtap and Not-Gate
|
Gibson-Assembly
1. Prepare 5X ISO buffer. Six ml of this buffer can be prepared by combining the following:
3 ml of 1 M Tris-HCl pH 7.5150 μl of 2 M MgCl260 μl of 100 mM dGTP 60 μl of 100 mM dATP 60 μl of 100 mM dTTP60 μl of 100 mM dCTP300 μl of 1 M DTT 1.5 g PEG-8000300 μl of 100 mM NADAdd water to 6 ml Aliquot 100 μl and store at -20 °C
2. Prepare an assembly master mixture. This can be prepared by combining the following:
320 μl 5X ISO buffer0.64 μl of 10 U/ μl T5 exo20 μl of 2 U/μl Phusion pol160 μl of 40 U/μl Taq ligAdd water to 1.2 ml
Aliquot 15 μl and store at -20 °C. This assembly mixture can be stored at -20 °C for at least one year. The enzymes remain active following at least 10 freeze-thaw cycles. This is ideal for the assembly of DNA molecules with 20-150 bp overlaps. For DNA molecules overlapping by larger than 150 bp, prepare the assembly mixture by using 3.2 μl of 10 U/ μl T5 exo.
3. Thaw a 15 μl assembly mixture aliquot and keep on ice until ready to be used.
4. Add 5 μl of DNA to be assembled to the master mixture. The DNA should be in equimolar amounts. Use 10-100 ng of each ~6 kb DNA fragment. For larger DNA segments, increasingly proportionate amounts of DNA should be added (e.g. 250 ng of each 150 kb DNA segment).
5. Incubate at 50 °C for 15 to 60 min (60 min is optimal).
6. If cloning is desired, electroporate 1 μl of the assembly reaction into 30 μl electrocompetent E. coli.
Documentation:
Why are you doing this experiment? Name the parts for the Gibson-Assembly.
| Label
| Concentrations ng/ μl
| μl to add to Gibson
|
| 4
| 55
| 0,5
|
| 5
| 40
| 0,6
|
| 6
| 50
| 0,5
|
| 9
| 47
| 0,55
|
| 10
| 56
| 0,45
|
| 11
| 30
| 0,8
|
4 with 9, 5 with 10, 6 with 11
|
Probes
|
| 2
| 42
| 4
|
| Inserts
| 4,9
| 5,10
| 6,11
|
| μl H2o
| 4
| 4
| 3,7
|
Describe your results and mistakes. Did you digest it? Results?
| See gel next day
>Did not work due to wrong DNA concentrations and no PCR purification
|
How did you label your samples and where are they stored?
Digestion
| Name:Rüdiger
| Date: 31.08.
|
| Continue from Date 31.08. Name Rüdiger
Experiment PCR
|
| Project Name: Precipitator
|
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng) (Vector:Insert ratio 1:3 in following Ligation)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample Name
| DNA concentration (μg/μl)
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Restriction enzymes you need to cut the vector, insert1 and insert 2:
| Components
| Vector (μl)
| Insert1 and 2 (μl)
|
| DNA (500ng)
|
|
|
| BSA (10x) (5μl)
|
|
|
| NEB4 Buffer (5μl)
|
|
|
| Enzyme 1 (1μl)
|
|
|
| Enzyme 2 (1μl)
|
|
|
| H2O (38 μl- DNA)
|
|
|
| In total 50 μl
|
|
|
Documentation:
| Name
| Vector (resistance)
(all +Antarctic phosphatase 1μl +AP Buffer 5,6μl )
| Insert
(all +Dpn1 1μl)
| Enzyme
| Enzyme 2
|
| 1V
| 1I
| PGEX (David)
| PCR29.08.-1
| EcoR1
| BamH1
|
| 2V
| 2I
| PGEX (David)
| PCR29.08.-2
| EcoR1
| BamH1
|
| 3V
| 3I
| PGEX (David)
| PCR29.08.-3
| EcoR1
| BamH1
|
| 4V
| 4I
| C3
| PCR29.08.-8
| EcoR1
| Spc1
|
| 5V
| 5I
| PET-DUET
| PCR31.08.-2a
| EcoR1
| PST1
|
| 6V
| 6I
| C3 (CM)
| PCR31.08.-42a
| EcoR1
| PST1
|
| 7V
| 7I
| C3 (CM)
| PCR31.08.-4a
| EcoR1
| PST1
|
| 8V
| 8I
| C3 (CM)
| PCR31.08.-2b
| EcoR1
| Spc1
|
| 9V
| 9I
| C3 (CM)
| PCR31.08.-42b
| EcoR1
| Spc1
|
| 10V
| 10I
| C3 (CM)
| PCR31.08.-4b
| EcoR1
| Spc1
|
|
Ligation
| Name: Sophie
| Date: 31.01.11
|
| Continue from Date: 30.08.11 Name: Sophie
Experiment: Digestion
|
| Project Name: inducible promoter for pbd
|
Procedure
PCR tube:
total volume 20 μl
- add H2O (17 μl -X-Y-Z)
- add 2 μl Ligase Buffer 10x
- add Insert 1, Insert 2(when proceeding from 3A digestion use 2 μl of each)
- add Vector (20ng needed. When proceeding from 3A digestion use 2 μl)
- Add 1 μl T4-DNA Ligase
- Incubate 10-30 min at room temperature
- heat for 20 minutes at 80°C
- store at -20°C or directly proceed to transformation
|
| Name of part
| Ratio Insert:Vector
= 3:1 or 1:1
| Volume (μl)
|
| X insert 1
| S54
| both
|
|
| Y insert 2
| ε 5
|
|
|
| Z vector
| psb1C3
|
|
|
| H2O
|
|
|
|
Documentation:
Why are you doing this experiment? Where are your parts stored? Name the parts for ligation etc.
Transformation
| Name:Sophie
| Date: 31.08.11
|
| Continue from Date: 31.08.11 Name: Sophie
Experiment: Ligation
|
| Project Name: inducible promoter for pbd
|
Procedure
- take cells from -80°C freezer and put them on ice! (every eppi contains about 400 μl cells)
- thaw cells on ice 20 minutes
- pipette 50 μl cells and 2 μl DNA into eppi still on ice!
- Incubate for 30 minutes on ice
- Heat at 42°C for 60 sec
- Incubate on ice for 5 minutes
- Add 200 μl LB Broth
- Incubate for 2 hours at 37°C (cells with lysis cassette at 30°C!!)
- Plate 50 μl and 200μl on two different LB/Agar plates with appropriate antibiotic resistance
Documentation:
Why are you doing this experiment? Name of the sample? Where are they stored? Name the vector with inserts, antibiotika resistance etc.
| Name: S54-ε 5-C3 1:1 / 1:3
stored in incubator on Cm plates
|
PCR
| Name: Sophie
| Date: 31.08.11
|
| Continue from Experiment new (Date)
(Name)
|
| Project Name:: pbd in Gst-vector
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| P93
|
| 2.5µl
| Primer dw
| P94
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
|
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
First 25 cycles touchdown 63°C -0,3°C per cycle
next 10 cycles touchdown 72°C -0,3°C per cycle
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
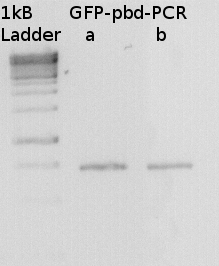
How did you label the PCR-Product, where is it stored and what do you do next?
Labeled GFP-pbd-PCR
stored in
I will digest it with Dpn I and with bamH I and Xho and ligate it to a GST-vector.
Digestion
| Name: Sophie
| Date: 31.08.11
|
| Continue from Date: 31.08.11 Name: Sophie
Experiment: PCR
|
| Project Name: pbd into GST-vector
|
Procedure
- add H2O (38μl-DNA )
- 5 μl NEB4 buffer (stored at iGEM’s, -20°C)
- 5 μl 10x BSA (used 1:10 diluted sample stored at iGEM’s, -20°C)
- DNA (500 ng) (Vector:Insert ratio 1:3 in following Ligation)
- 1 μl restriction enzymes (stored at iGEM’s, -20°C)
- heat for 1-2 hours 37°C (6 hours if time)
- add 5,6 μl phosphatase buffer and 1 μl antarctic phosphatse to the vector and digest for another hour
- heat for 20 minutes 80°C (inactivation of enzymes)
- keep at 4°C if you cannot continue
Measured DNA-concentration with Nanodrop to calculate the volume of DNA to do the digestion:
| Sample Name
| DNA concentration (μg/μl)
|
| pGex
| 480
|
| gFP-pbd-PCR
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Restriction enzymes you need to cut the vector, insert1 and insert 2:
| Components
| Vector (μl)
| Insert
|
| DNA (500ng)
|
|
|
| BSA (10x) (5μl)
|
|
|
| NEB4 Buffer (5μl)
|
|
|
| Enzyme 1 (1μl)
| BamH I
| BamH I
|
| Enzyme 2 (1μl)
| Xho I
| Xho I
|
| H2O (38 μl- DNA)
|
|
|
| In total 50 μl
|
|
|
Documentation:
Why are you doing this experiment? Where are the samples stored? Antibiotica resistance, vector used etc.
| Why? I want to produce pbd-protein to make experiments and to get data for the modeling.
Vector: pGEX from Hemin
insert: GFP-pbd-PCR
|
 "
"
 Contact
Contact 












