Team:TU Munich/lab/notebook/part2
From 2011.igem.org
| Line 4: | Line 4: | ||
<div class="ui-corner-all subcontent tabcontent"> | <div class="ui-corner-all subcontent tabcontent"> | ||
| + | <table id="toc" class="toc"><tr><td><div id="toctitle"><h2>Contents</h2></div> | ||
| + | <ul> | ||
| + | <li class="toclevel-1 tocsection-1"><a href="#Light_sensor_systems_and_AND-Gate_cloning_Part_II_Alex.2FSusan"><span class="tocnumber">1</span> <span class="toctext">Light sensor systems and AND-Gate cloning Part II Alex/Susan</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-2 tocsection-2"><a href="#09-08-2011"><span class="tocnumber">1.1</span> <span class="toctext">09-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-3"><a href="#Cloning"><span class="tocnumber">1.1.1</span> <span class="toctext">Cloning</span></a> | ||
| + | |||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-4"><a href="#Digestions"><span class="tocnumber">1.1.1.1</span> <span class="toctext">Digestions</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-5"><a href="#Other_Work"><span class="tocnumber">1.1.2</span> <span class="toctext">Other Work</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-6"><a href="#10-08-2011"><span class="tocnumber">1.2</span> <span class="toctext">10-08-2011</span></a> | ||
| + | |||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-7"><a href="#Cloning_2"><span class="tocnumber">1.2.1</span> <span class="toctext">Cloning</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-8"><a href="#Digestion"><span class="tocnumber">1.2.1.1</span> <span class="toctext">Digestion</span></a></li> | ||
| + | <li class="toclevel-4 tocsection-9"><a href="#Transformation"><span class="tocnumber">1.2.1.2</span> <span class="toctext">Transformation</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-10"><a href="#Other_Work_2"><span class="tocnumber">1.2.2</span> <span class="toctext">Other Work</span></a></li> | ||
| + | |||
| + | <li class="toclevel-3 tocsection-11"><a href="#Results"><span class="tocnumber">1.2.3</span> <span class="toctext">Results</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-12"><a href="#11-08-2011"><span class="tocnumber">1.3</span> <span class="toctext">11-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-13"><a href="#Other_Work_3"><span class="tocnumber">1.3.1</span> <span class="toctext">Other Work</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-14"><a href="#Testing"><span class="tocnumber">1.3.2</span> <span class="toctext">Testing</span></a></li> | ||
| + | |||
| + | <li class="toclevel-3 tocsection-15"><a href="#Cloning_3"><span class="tocnumber">1.3.3</span> <span class="toctext">Cloning</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-16"><a href="#Ligation_and_Transformation"><span class="tocnumber">1.3.3.1</span> <span class="toctext">Ligation and Transformation</span></a></li> | ||
| + | <li class="toclevel-4 tocsection-17"><a href="#Digestion_2"><span class="tocnumber">1.3.3.2</span> <span class="toctext">Digestion</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-18"><a href="#Results_2"><span class="tocnumber">1.3.4</span> <span class="toctext">Results</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-19"><a href="#12-08-2011"><span class="tocnumber">1.4</span> <span class="toctext">12-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-20"><a href="#Other_Work_4"><span class="tocnumber">1.4.1</span> <span class="toctext">Other Work</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-21"><a href="#Cloning_4"><span class="tocnumber">1.4.2</span> <span class="toctext">Cloning</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-22"><a href="#Digestion_3"><span class="tocnumber">1.4.2.1</span> <span class="toctext">Digestion</span></a></li> | ||
| + | |||
| + | <li class="toclevel-4 tocsection-23"><a href="#Ligation_and_Transformation_2"><span class="tocnumber">1.4.2.2</span> <span class="toctext">Ligation and Transformation</span></a></li> | ||
| + | <li class="toclevel-4 tocsection-24"><a href="#Transformation_2"><span class="tocnumber">1.4.2.3</span> <span class="toctext">Transformation</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-25"><a href="#Testing_2"><span class="tocnumber">1.4.3</span> <span class="toctext">Testing</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-26"><a href="#Results_3"><span class="tocnumber">1.4.4</span> <span class="toctext">Results</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-27"><a href="#16-08-2011"><span class="tocnumber">1.5</span> <span class="toctext">16-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-28"><a href="#Cloning_5"><span class="tocnumber">1.5.1</span> <span class="toctext">Cloning</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-29"><a href="#Transformation_3"><span class="tocnumber">1.5.1.1</span> <span class="toctext">Transformation</span></a></li> | ||
| + | </ul> | ||
| + | |||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-30"><a href="#Other_Work_5"><span class="tocnumber">1.5.2</span> <span class="toctext">Other Work</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-31"><a href="#Results_4"><span class="tocnumber">1.5.3</span> <span class="toctext">Results</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-32"><a href="#17-08-2011"><span class="tocnumber">1.6</span> <span class="toctext">17-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-33"><a href="#Cloning_6"><span class="tocnumber">1.6.1</span> <span class="toctext">Cloning</span></a> | ||
| + | |||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-34"><a href="#MiniPrep_and_Digestion"><span class="tocnumber">1.6.1.1</span> <span class="toctext">MiniPrep and Digestion</span></a></li> | ||
| + | <li class="toclevel-4 tocsection-35"><a href="#Transformation_4"><span class="tocnumber">1.6.1.2</span> <span class="toctext">Transformation</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-36"><a href="#Testing_3"><span class="tocnumber">1.6.2</span> <span class="toctext">Testing</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-37"><a href="#Results_5"><span class="tocnumber">1.6.3</span> <span class="toctext">Results</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-38"><a href="#18-08-2011"><span class="tocnumber">1.7</span> <span class="toctext">18-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-39"><a href="#Other_Work_6"><span class="tocnumber">1.7.1</span> <span class="toctext">Other Work</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-40"><a href="#Results_6"><span class="tocnumber">1.7.2</span> <span class="toctext">Results</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | |||
| + | <li class="toclevel-2 tocsection-41"><a href="#19-08-2011"><span class="tocnumber">1.8</span> <span class="toctext">19-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-42"><a href="#Other_Work_7"><span class="tocnumber">1.8.1</span> <span class="toctext">Other Work</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-43"><a href="#Results_7"><span class="tocnumber">1.8.2</span> <span class="toctext">Results</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-44"><a href="#22-08-2011"><span class="tocnumber">1.9</span> <span class="toctext">22-08-2011</span></a> | ||
| + | |||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-45"><a href="#Results_8"><span class="tocnumber">1.9.1</span> <span class="toctext">Results</span></a></li> | ||
| + | <li class="toclevel-3 tocsection-46"><a href="#Cloning_Blue_Light_Sensor"><span class="tocnumber">1.9.2</span> <span class="toctext">Cloning Blue Light Sensor</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-47"><a href="#Digestion_4"><span class="tocnumber">1.9.2.1</span> <span class="toctext">Digestion</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-48"><a href="#Other_Work_8"><span class="tocnumber">1.9.3</span> <span class="toctext">Other Work</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-2 tocsection-49"><a href="#23-08-2011"><span class="tocnumber">1.10</span> <span class="toctext">23-08-2011</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-3 tocsection-50"><a href="#Cloning_Blue_Light_Sensor_2"><span class="tocnumber">1.10.1</span> <span class="toctext">Cloning Blue Light Sensor</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-51"><a href="#Digestion_5"><span class="tocnumber">1.10.1.1</span> <span class="toctext">Digestion</span></a></li> | ||
| + | <li class="toclevel-4 tocsection-52"><a href="#Transformation_5"><span class="tocnumber">1.10.1.2</span> <span class="toctext">Transformation</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-53"><a href="#Testing_4"><span class="tocnumber">1.10.2</span> <span class="toctext">Testing</span></a> | ||
| + | <ul> | ||
| + | <li class="toclevel-4 tocsection-54"><a href="#In_Vitro_Assay"><span class="tocnumber">1.10.2.1</span> <span class="toctext">In Vitro Assay</span></a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li class="toclevel-3 tocsection-55"><a href="#Results_9"><span class="tocnumber">1.10.3</span> <span class="toctext">Results</span></a></li> | ||
| + | |||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | </td></tr></table><script>if (window.showTocToggle) { var tocShowText = "show"; var tocHideText = "hide"; showTocToggle(); } </script> | ||
| + | |||
<h1> <span class="mw-headline" id="LightsensorsystemsandAND-Gatecloning.2FSusan">Cloning Part II</span></h1> | <h1> <span class="mw-headline" id="LightsensorsystemsandAND-Gatecloning.2FSusan">Cloning Part II</span></h1> | ||
<b>People: Alex H., Susan</b> | <b>People: Alex H., Susan</b> | ||
Revision as of 18:44, 21 September 2011
Contents[hide] |
Cloning Part II
People: Alex H., Susan09-08-2011
People: Alex H. and Susan
Cloning
Digestions
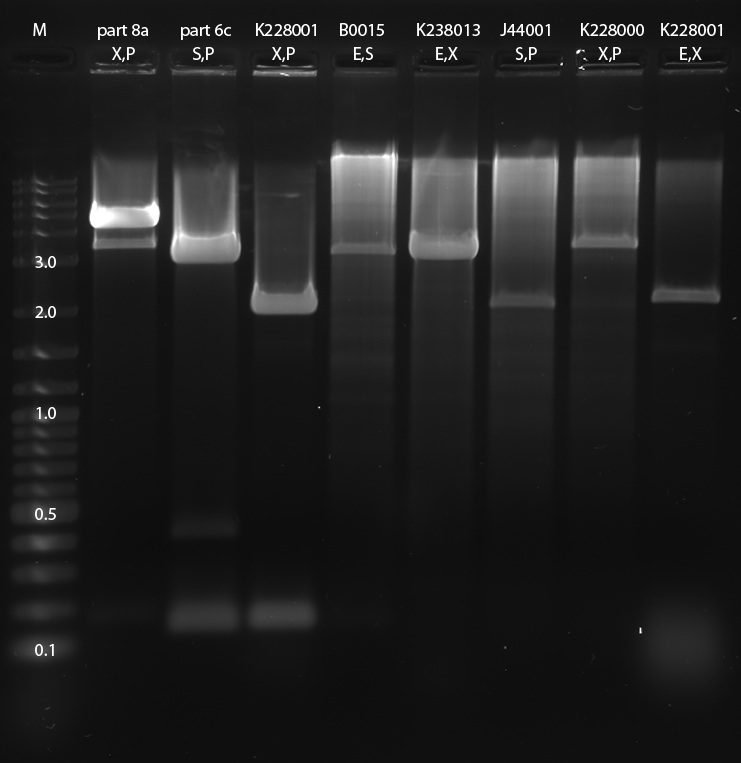
Restriction digests: part 8a (RBS-T7Pol) (X,P), part 6c (Terminator-BlueLight) (S,P), SupDtRNA (X,P), B0015 (E,S), K238013 (E,X), J44001 (S,P), K228000 (X,P) SupDtRNA (E,X) parts 8a and 6c might be switched, we will confirm it on the gel tomorrow), SupDtRNA was restricted twice in order to try out different ligation strategies
Due to power cut tomorrow no overnight cultures can be done
Other Work
Sequencing of following parts was done by Kathi: T7promotor-lacZ (I712074-I732017), Terminator-blue light sensor (B0015-K238013), T7 Pol-RBS (K228000-J44001) --> Results: not clear! smaller parts seem to be missing in the sequence (but: these parts were already on the plasmid which were used for ligation)
THEREFORE: try again!!! and repeat ligations from 21/7 and 12/7
10-08-2011
People: Alex H. and Susan
Cloning
Digestion
Gel electrophoresis: An Agarose gel electrophoresis (1%, 1x TAE buffer) of the eight digested samples from yesterday was performed (running time approximately 1.5 h).
Transformation
Transformation: In order to yield further DNA of part 2b (lacZ-T7 promotor), 1 ul of part 2b was mixed with 40 ul electrocompetent BL21 bacteria. Transformation was done according to the protocol. After incubation for 2 h in SOC-medium, bacteria were plated onto Amp-plates and incubated at 37 °C over night.
Other Work
Test of electrocompetent cells: DH5a cells were inoculated on LB plates (A, K or Cm) and incubated over night at 37 °C.
Results
Gel electrophoresis: We expect the following bands to appear on the gel: part 8a (X,P): ca. 2.7 kbp (insert: T7 pol-rbs) and ca. 2 kbp (backbone) part 6c (S,P): ca. 2.2 kbp (backbone including blue light promotor-terminator) SupDtRNA (X,P): ca. 0.15 kbp (insert: SupDtRNA) and ca. 2 kbp (backbone) B0015 (E,S): ca. 0.15 kbp (insert: terminator) and ca. 3.1 kbp (backbone) K238013 (E,X): ca. 2.1 kbp (backbone including K238013) J44001 (S,P): ca. 2.1 kbp (backbone including J4401) K228000 (X,P): ca. 2.7 kbp (K228000) and ca. 2 kbp (backbone) SupDtRNA (E,X): ca. 2.1 kbp (backbone including SupDtRNA)
If 6c and 8a were exchanged by mistake (see above), the following bands should appear: part 8a (S,P): ca. 4.7 kbp (backbone including T7 pol-rbs) part 6c (X,P): ca. 0.2 kbp (insert: blue light promotor-terminator) and ca. 2 kbp (backbone)
The picture of the gel was named 100811GelVerdau0809SusanAlex. Digestion was successful for SupDtRNA (X,P), SupDtRNA (E,X), J4401 (S,P) and K228000 (X,P). Digestion of K238013 (E,X) yielded a band of about 2.7 kbp, which contradicts our expectations. part 8a and part 6c were exchanged by mistake, as they show the bp calculated for the exchanged parts. DNA was extracted using freeze and squeeze.
11-08-2011
People: Susan and Alex
Other Work
Gene synthesis: We inquired Invitrogen and ShineGene for possible gene synthesis of our plasmids, in case we do not make it in time to synthesize them.
New electrocompetent cells: We inoculated new DH5a cells in 20 ml LURIA medium, as our own ones seem to be contaminated.
Test of electrocompetent cells: Miniprep with Metabion Kit of DH5a cells in LB (overnight culture from 10-8-2011)
Testing
Test of lacZ expression in plasmid 2 with T7 promotor (miniprep 2b from 14-7-2011): colony picking and growth in 5 ml LB-medium with 5ul Amp (Start 9:45h) OD measurement: at 16:00h finally 0.775 IPTG induction: 40ul IPTG (0.1M) in 2ml cells (since cells didn't grow as fast as expected experiment is repeated tomorrow) --> 4 new cultures are raised overnight in 5ml LB with Amp (two from 100ul Transformation culture, 2 from 100ul concentrated culture)
Before induction, cells were spread on two amp-LB plates: negative control with 16ul (should be 20mg/ml) SGal (50mg/ml), and test for lacZ expression with 16ul SGal and 40ul (IPTG (0.1M in H2O)
Cloning
Ligation and Transformation
Ligation of the digested parts from yesterday: J44001 (rbs) with K228000 (T7 Pol) and K322127 (red light sensor) with K228001 (tRNA). After ligation, the plasmids were transformed by electroporation into competent DH5alpha E. coli (by Andrea, not our own stock).
Digestion
Repetition of the digestions from yesterday for B0015 (E,S; double terminator; was conducted with two different lots) and K238013 (E,X; blue light promotor).
Results
Test of electrocompetent cells: Colonies have appeared on ampicillin, kanamycin and chloramphenicol plates. Thus, our DH5a stock is most likely contaminated...
12-08-2011
People: Susan and Alex
Other Work
The preparation of new electrocompetent DH5alpha cells (from Andrea) is continued. The cells yielded only a low optical density and therefore have to be tested on functionality.
Cloning
Digestion
Gel electrophoresis for digestion and contamination test from yesterday: The digested samples (2x B0015, K238013) were applied to a 1 % gel. The prep from our (contaminated) old DH5alpha cells was also applied to the same gel. The gel was run at 90 V for ca. 50 min. The lanes are as follows (from left to right): 2log DNA ladder (5 ul); B0015 digestion 1 (20 ul); B0015 digestion 1 (20 ul); B0015 digestion 2 (20 ul); B0015 digestion 2 (20 ul); K238013 digestion (20 ul); K238013 digestion (20 ul); DH5alpha contamination test prep (10 ul)
Ligation and Transformation
Ligation and transformation of B0015 and K238013: The digested parts (B0015 and K238013) were cut from the gel and the DNA was extracted using freeze 'n squeeze. For B0015, DNA from digestion 2 (lanes 4 and 5) were used. For K238013, two bands at approximately 2.7 kbp were extracted (an upper and a lower band). Ligation was performed with DNA extracted from the upper or lower band, and B0015. The ligated plasmid was transformed into DH5alpha cells (from Andrea), which were plated onto LB with Chloramphenicol. The plates were incubated for one day at 37 °C and after that, at 4 °C until Tuesday.
Transformation
Transformation of part 2b (T7 prom-lacZ): Part 2b was transformed by electroporation into DH5alpha cells (from Andrea). They were plated onto LB(Kan) plates. This is in order to have a non-contaminated DH5alpha strain containing part 2b.
Testing
Expressionstest 2 - SDS-PAGE: 4 overnight cultures of T7-lacZ in BL21 Zellen (see yesterday): OD Measurement: 1: to low, 2: 1.2 (from unconcentrated plate), 3+4: 1.0 (concentrated plate) --> 1.5ml of culture 2 and 3 were raised in 3ml new LB with Amp for one hour. OD = ca. 0.7 --> induction with IPTG (endconcentration 1mM). 500ul samples are taken at: start, 1h, 2h, 3h, 4h 250ul of each sample were spun down, resuspended in 50ul H20 and 10ul Laemmli buffer (kindly provided by Evi) and incubated at 95°C for 5min. Hopefully this should lead to cell lysis. 20ul of samples were loaded on a 10% SDS Page (protocol from Evi). Protein ladder: PageRuler™ Plus Prestained Protein Ladder by Thermofisher.
Results
Transformation: The transformations of J44001 (rbs) with K228000 (T7 Pol) and K322127 (red light sensor) with K228001 (tRNA) did notyield any colonies on the plates. We have to repeat this.
LacZ expression test: The cells plated onto IPTG and S-Gal did not yield black colonies. This might be evidence for a malfunction of our 2b part... We will test this again today by a SDS-PAGE for overexpressed LacZ.
Contamination tests: It seems that all our electrocompetent cells (DH5alpha, BL21 and BM28) seem to be contaminated. Colonies grew on Amp (not BL21), Kan (not BL21) and Cm plates. Electrocompetent cells from Andrea were also plated as negative controls, they did not yield colonies (or only very few ones).
Expressionstest 2 - SDS-PAGE:
An increase in the intensity of the 10 kDa band would have been expected, caused by elevated expression levels of lacZ alpha peptide.
16-08-2011
People: Alex
Cloning
Transformation
Transformation of part 2b into BL21: Part 2b was transformed by electroporation into BL21 cells (from Andrea). They were plated onto LB(Kan) plates. This is in order to repeat the lacZ expression test from 11-08-2011.
Other Work
New Medium and Plates: New LB medium and agar plates containing Amp, Kan and Cm were prepared. New 10% Glycerol was prepared.
Results
Transformations: Many colonies grew on the plates containing DH5alpha cells transformed with part 2b. Only very few colonies grew on the plates containing DH5alpha cells transformed with ligation of B0015 and K238013 -> (most likely) non at all on the plates from the ligation with the lower band of K238013; all colonies on the plates of the ligation with the upper band of K238013 are pink (= unsuccessful digestion and ligation?!?). There was one white colony, which was inoculated.
Contamination test of new electrocompetent cells: No colonies grew on the plates inoculated with the freshly produced electrocompetent cells (A, K or Cm).
17-08-2011
People: Alex
Cloning
MiniPrep and Digestion
Mini-Prep and control digestion: Plasmids of cultures of 5 colonies of part 2b and 2 colonies (1 white, other one maybe white) of ligation of blue light promotor and double terminator. The plasmids of 2 cultures each are digest by EcoR1 and Spe1. They are applied, together with undigested plasmids, onto a 1 % Agarose gel. The gel was run for ca. 1:30 h at 90 V. The lanes are as follows (from left to right): 2log DNA ladder (3 ul); part 2b Nr. 1 (20 ul); part 2b Nr. 1 digested (20 ul); part 2b Nr. 2 (20 ul); part 2b Nr. 2 digested (20 ul); ligation of blue light promotor & terminator Nr. 1 (20 ul); ligation of blue light promotor & terminator Nr. 1 digested (20 ul); ligation of blue light promotor & terminator Nr. 2 (20 ul); ligation of blue light promotor & terminator Nr. 2 digested (20 ul).
Transformation
Transformation of K322127: The part K322127 is transformed into BL21 (from Andrea) and DH5alpha (our new stock). The BL21 culture should be used in order to further characterize the sensor, while the DH5aplha transformation is in order to test the freshly prepared electrocompetent cells. The cells are plated onto LB (Cm) plates.
Testing
Expression-test of part 2b in BL21: Four colonies of the successful transformations are inoculated in LB applied with Kan and incubated for several hours. At an approximate OD = 0.77, they are plated onto LB plates (Kan), supplied with 16 ul S-Gal (50 mg/ml) and 40 ul IPTG (0.1 M), or only S-Gal. The plates are incubated over night in aluminium foil, in order to ensure darkness. Prior to plating of the bacteria, two over night cultures (5 ml LB with Kan) were inoculated with cultures Nr. 3 and 4. They are used on the next day to perform an expression test by SDS-PAGE analysis.
Results
Transformation of 2b in BL21: Many colonies on both plates (normal and concentrated).
Plasmid concentrations: Plasmid concentrations were measured on Nanodrop, using 3 ul sample. Part 2b Nr. 1 -> 109 ng/ul; Part 2b Nr. 2 -> 121 ng/ul; Part 2b Nr. 3 -> 106 ng/ul; Part 2b Nr. 4 -> 130 ng/ul; Part 2b Nr. 5 -> 118 ng/ul; Ligation blue light prom & terminator Nr. 1 -> 89 ng/ul; Ligation blue light prom & terminator Nr. 2 -> 68 ng/ul.
18-08-2011
People: Alex
Other Work
Primer Design: New primers for sequencing of blue light promotor & double terminator (K238013 & B0015), T7 Pol & rbs (K228000 & J44001) and lacZ & T7 promotor were generated. Sequencing will be done tomorrow, as the necessary bar codes are not available, yet.
Preparation for new expression tests: New liquid cultures (5 ml LB with Kan) are inoculated with BL21 cells transformed with part 2b (T7 promotor & lacZ). One hour after measurement of OD between 0.58-0.62 they are induced with 1 mM IPTG (control: no induction) and incubated over night. The experiment is prepared in two sets: one for expression test by SDS-PAGE, the other one by Miller-Assay.
Preparation for expression test with red light sensor: Liquid cultures (5 ml LB with Cm) are inoculated with BL21 cells transformed with K322127. They are incubated in the box supplied with the 628 nm diode and incubated over night. The following day, S-Gal expression tests will be conducted.
Results
Expression test of lacZ: The colonies on the S-Gal plates supplied with IPTG again did NOT yield black coloring. This experiment will be repeated tomorrow in liquid culture with cells induced over night.
Testing of new electrocompetent cells: The freshly prepared electrocompetent cells seem to work. However, they yield much less transformants than usual.
19-08-2011
People: Alex
Other Work
Sequencing: Part 2b, 6c and 8a, as well as a new ligation of K238013 and B0015 are sent to sequencing by GATC.
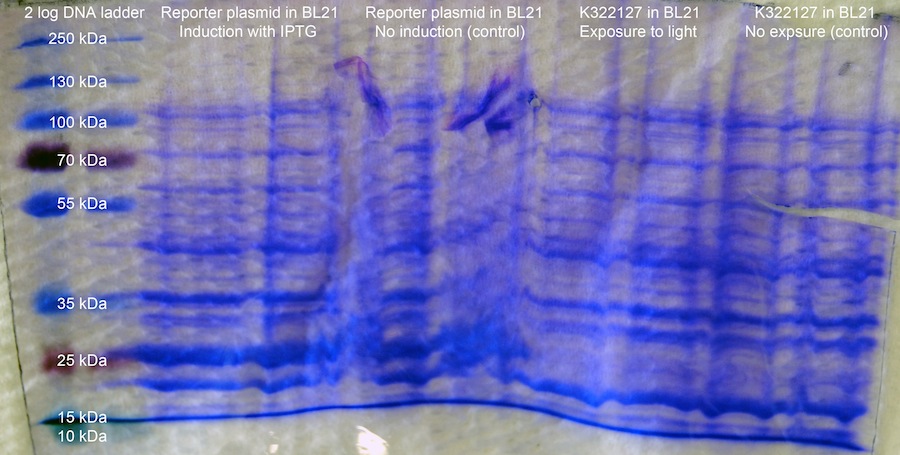
Expression analysis - SDS-PAGE: A 10 % SDS-PAGE is prepared. The cultures of K322127 in BL21 exposed to the red light diode are exposed for three hours to the light of a desk lamp, or are continually exposed to the red light (control). The OD of the over night cultures from 18-08-2011 (part 2b in BL21 and K223217 in BL21) are measured. 1 OD is centrifuged and the cells are resuspended in 50 ul Laemmli-Buffer. The samples are boiled for 5 min and 15 ul are supplied onto the gel in the following order: 2x 2log DNA ladder; 2x reporter plasmid in BL21, induced by IPTG; 2x Reporter Plasmid in BL21, not induced; 2x K322127 in BL21, induced by light; 2x K322127 in BL21, not induced. The gel is run for 1:30 h at 80 V, which was increased to 12 V, as soon as the buffer front reached the separation gel.
Expression analysis - S-Gal Assay: 0.5 OD from the samples metioned above are filled into cuvettes. S-Gal and FAS are added and the OD spectra are measured every 30 seconds.
Results
b-galactosidase expression assay by S-Gal: The assay did not yield the desired results, as there were problems with the OD of S-Gal. S-Gal itself seems to absorb very strongly, which interfered with the measurement in the spectrometer. Furthermore, the culture was visibly darkening in both the sample and the negative control, which should not have been the case.
Expression Assay by SDS-PAGE:
The gel cannot be discussed properly. Most likely, it took damage while being destained. It is also possible that the applied voltage was too high. A difference in band intensity at approximately 10 kDa, which is the size of the lacZ alpha peptide, would have been expected.
22-08-2011
People: Alex, Simon
Results
Sequencing: Some of the sequenced data arrived. Part 8a (from 14-07-2011; T7Pol & rbs) does NOT contain the desired rbs. The T7Pol seems to be inserted alright. For the rest of the data, GATC awaits the arrival of the necessary primers.
Cloning
Digestion
-part "T7" (date: 17.6., concentration = 190 ng/µl) was digested using X and P. At first we thought that this is a T7 polymerase part (the tube wasn't properly labeled!!!), but later we found out that this is really a T7 promoter.
- part "RBS minip" (date: 22.6., concentration = 47.5 ng/µl) was digested using S and P.
- part "K238013 cm" (date: 29.6., concentration = 233 ng/µl) was digested using S and P.
Analytic & preparative Gel:
The samples from the restriction digests (see above) were separated using a 1% Agarosegel. Expectations: T7prom cut with X,P should yield a band at approximately 3.5 kbp. Rbs cut with S,P should yield a band at approximately 2 kbp. K238013 in pSB3C5 should yield a band at approximately 2.8 kbp. In pSB1A2 it should be 2 kbp.
The original photography cannot be shown, as the file seems to be corrupt. The bands all fit, except for K238013. The content of this plasmid seems unclear, as the length in base pairs is much higher than expected. For the upcoming ligation, another K238013 should be used.
Other Work
Inoculation: 5 ml LB (Kan) were inoculated with two further colonies from the transformations of the reporter plasmid (part2b) into BL21 from DATUM. The cultures were grown over night.
LB S-Gal Paltes: S-Gal plates were produced, using 500 ml ddH2O, 12.5 mg LB, 300 mg S-Gal and 460 mg ferric ammonium sulfate. After autoclavation, the solutions were tempered to about 58 °C and Kan or Cm was added. The solutions were spilled into plates and are kept in the fridge.
23-08-2011
People: Alex, Simon, Bea, Thorsten
Cloning
Digestion
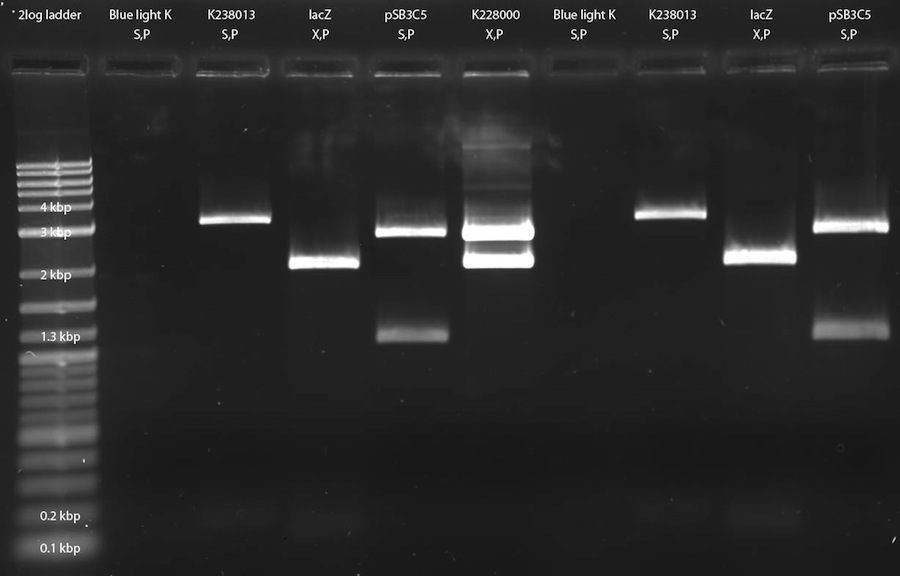
We digested blue light K from 29-06-2011 (S,P), K238013 in unknown plasmid (S,P), lacZ (X,P) and pSB3C5 (S,P). After digestion, the samples were applied onto a 1 % gel. The lanes are as follows (from left to right): 2log DNA ladder (3 ul); lacZ (20 ul); blue light K (20 ul); K238013 (20 ul); pSB3C5 (20 ul); K228000 (20 ul); lacZ (20 ul); blue light K (20 ul); K238013 (20 ul); pSB3C5 (20 ul). LacZ did not show any bands and blue light K yielded a band at an unexpected heigth, but the rest of the bands are as expected.
The following parts were extracted and frozen: Blue light K at ca. 3 kbp; K238013 at ca. 2.2 kbp; pSB3C5 at ca. 2.7 kbp; T7pol at ca. 2.7 kbp and the plasmid of T7pol (pSB1A2) at ca. 2 kbp. They will be used for ligation tomorrow.
Transformation
Blue light K from 29-06-2011 was transformed into DH5alpha cells and plated onto LB (Cm) plates, which were incubated over night.
Induction of reporter plasmid: The OD was measured from the LB (Kan) cultures inoculated with the reporter plasmid (part2b) in BL21, which were incubated over night. At an OD of about 0.7, the cultures were split and one half of the cultures were induced with IPTG (cfinal = 1mM). After a few hours, the cultures were plated onto S-Gal plates with Kann and incubated over night.
Testing
Test of K322127: 5 ml LB with Cm were inoculated each with DH5alpha transformed with K322127. The cultures are incubated over night during exposition to a LED with approximately 630 nm.
In Vitro Assay
In Vitro Assay (Bea, Simon):
To assess the reporter plasmid ("Part 2b"). According to the DNA Sequence (see meetings 22.08. and 15.8) the part contains lacZ, but the sequence data is bad in the region of the T7 promotor. To find out whether the T7 promotor is present, we performed an in vitro assay. At first, the DNA samples of interest were transcribed using a T7 polymerase (for exact composition of reaction mixture see below), incubation time: 2h at 37 °C. After this, each sample was split in two halves. One of these each of these halves was then incubated at 37 °C for 30 minutes in presence of RNase. Finally, the different samples were separated in a 2% Agarose gel. The RNase should destroy the RNA produced by the T7 polymerase. Thus if two corresponding lines of the gel are compared, bands that only appear in one of the two lines are caused by RNA. This means that the respective DNA construct contained a T7 promoter. For exact protocol also see Methods.
The following DNA samples were transcribed in vitro using a T7 polymerase: Sample Number - Part Name - Date - concentration (ng/ul)
- 1 - 2b - 13.07.2011 - 215
- 2 - Trafo 2b Nr. 1 - 17.08.2011 - 106
- 3 - Trafo 2b Nr. 2 - 17.08.2011 - 121
- 4 - mistake - was thrown away
- 5 - Trafo 2b Nr. 4 - 17.08.2011 - 130
- 6 - Trafo 2b Nr. 5 - 17.08.2011 - 118
- Positive Control (PK) - Miniprep iGEM T7 - 31.05.11 - 58
The size of the construct in samples 1 - 6 is approx. 6.6 kb, the size of the construct of the positive Control is approx. 3.4 kb. Thus, the number of plasmids per microliter is approximately the same in all samples (only half the amount of sample 1 was used).
The reaction mixture contained: 2 µl buffer (40 mM TrisHCl, 40 mM MgCl2), 2 µl DTT (10 mM), 2 µl T7 Polymerase (50 u /µl), 10 µl DNA (only 5 µl in Sample 1), 1µl rNTPs (4 mM), fill up to 20 µl with ddH2O.
After incubation (2h at 37 °C), 10 µl of each sample was taken and mixed with 1 µl of RNase and incubated at 37 °C for 30 minutes. After this, all 12 samples were separated using a 2 % Agarose Gel (2log DNA ladder). Gel electrophoresis, 100 V, 400 mA, ca. 1:30 h.
Results
Transformations: The transformation of K238013 into DH5alpha cells on LB with Cm from yesterday was successful. However, all the colonies were pink (part not inserted!?). The T7promotor grew on Amp and Kan. From the last days: The transformation of B0015 and K238013 yielded pink bands (part not inserted!?).
In Vitro Assay
Samples #2 through #6a look very much alike, with the only difference being the "blurry blobs" at the bottom of the gel (not visible in #2a). We assume that these "blobs" are caused by RNA fragments, which leads us to the conclusion that RNase was present in all of these samples. The reason for this might be the fact that these samples were prepared from cells without performing an ethanol precipitation and thus, RNase from the cells might contaminate the samples. Looking at the samples #5 and #5a as well as #6 and #6a, for example, there is a band at about 6 kb. This must be DNA. This and the fact that there is digested RNA visible at the bottom leads us to the conclusion, that the band at 6 kb is a plasmid containing a T7 promoter (--> accordingly, this should be the correct plasmid containing T7 promoter and lacZ). To verify this, a second in vitro assay should be performed after inhibition of RNase in these samples. Comparison of #1 and #1a shows that part 2b must contain a T7 promoter, because of the blurred signal in #1 which is missing in #1b. This must be RNA which was digested by the RNase present in #1b. In both #1 and #1a there are bands at approximately 6 kb and 3 kb (6 kb could be the backbone containing T7 promoter and lacZ, 3 kb could be backbone containing only the t7 promoter). This DNA might be two different plasmids. This explains why the sequencing results are bad. At least one of these plasmids contains a T7 promoter, but it is unclear which one.
 "
"