Team:Potsdam Bioware/Methods
From 2011.igem.org
KatharinaB (Talk | contribs) (→Methods) |
KatharinaB (Talk | contribs) (→Methods) |
||
| Line 6: | Line 6: | ||
<br> | <br> | ||
| + | '''Cloning'''<br> | ||
| + | <br> | ||
| + | '''Polymerase Chain Reaction'''<br> | ||
| + | <br> | ||
| + | The purpose of PCR is to amplify a gene to get a huge number of copies. The sample is denaturated at 94°C to melt the double stranded DNA to single stranded. This will allow the primers to bind at a specific annealing temperature to the their complementary strand. Due to that the polymerase can start elongating the primer and copying the template. | ||
| + | |||
| + | '''PCR purification'''<br> | ||
| + | <br> | ||
| + | If the PCR conditions are well optimized, most of the added primers and dNTPs will be used up during the PCR amplification. For further cloning steps you need to have only one major PCR band visible on your agarose gel. To get rid of interfering factors you need to purify the PCR product. For purification we used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren) as well as the Wizard® SV Gel and PCR Clean-Up System from Promega (Darmstadt). | ||
Revision as of 14:48, 21 September 2011
Methods
Established Methods
Cloning
Polymerase Chain Reaction
The purpose of PCR is to amplify a gene to get a huge number of copies. The sample is denaturated at 94°C to melt the double stranded DNA to single stranded. This will allow the primers to bind at a specific annealing temperature to the their complementary strand. Due to that the polymerase can start elongating the primer and copying the template.
PCR purification
If the PCR conditions are well optimized, most of the added primers and dNTPs will be used up during the PCR amplification. For further cloning steps you need to have only one major PCR band visible on your agarose gel. To get rid of interfering factors you need to purify the PCR product. For purification we used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren) as well as the Wizard® SV Gel and PCR Clean-Up System from Promega (Darmstadt).
Cloning of BioBricks
All the genes for BioBrick production were amplified by PCR. Due to the incorporation of the sequences of the iGEM restriction enzymes into the primer sequence our biobricks comply with the BioBrick standards. We used the E. coli strain XL1 blue as host for cloning of all parts. The sequence of the final constructed BioBrick was verified by sequencing.
HPLC analysis
HPLC is used in analysis for identifying and purifying individual compounds in a mixture. We wanted to purify the microviridin for further analysis. After incubation of an E. coli culture for reaching an OD600 between 0.5 and 0.6 the cells were centrifuged and the pellet was washed with Tris-HCl. The pellet was resuspended in H2O. The cells were then lysed via sonication and centrifuged. The supernatant was transferred to Sep-Pak Plus C18 Cartridges. After washing the samples were eluted from the cartridge in methanol and put in a speedvac. Afterwards the remaining pellet was resuspended in methanol, filtered and put on a HPLC vial. HPLC analyses were carried out on a Shimadzu HPLC. Fractions were sampled by hand.
Principle of phage display
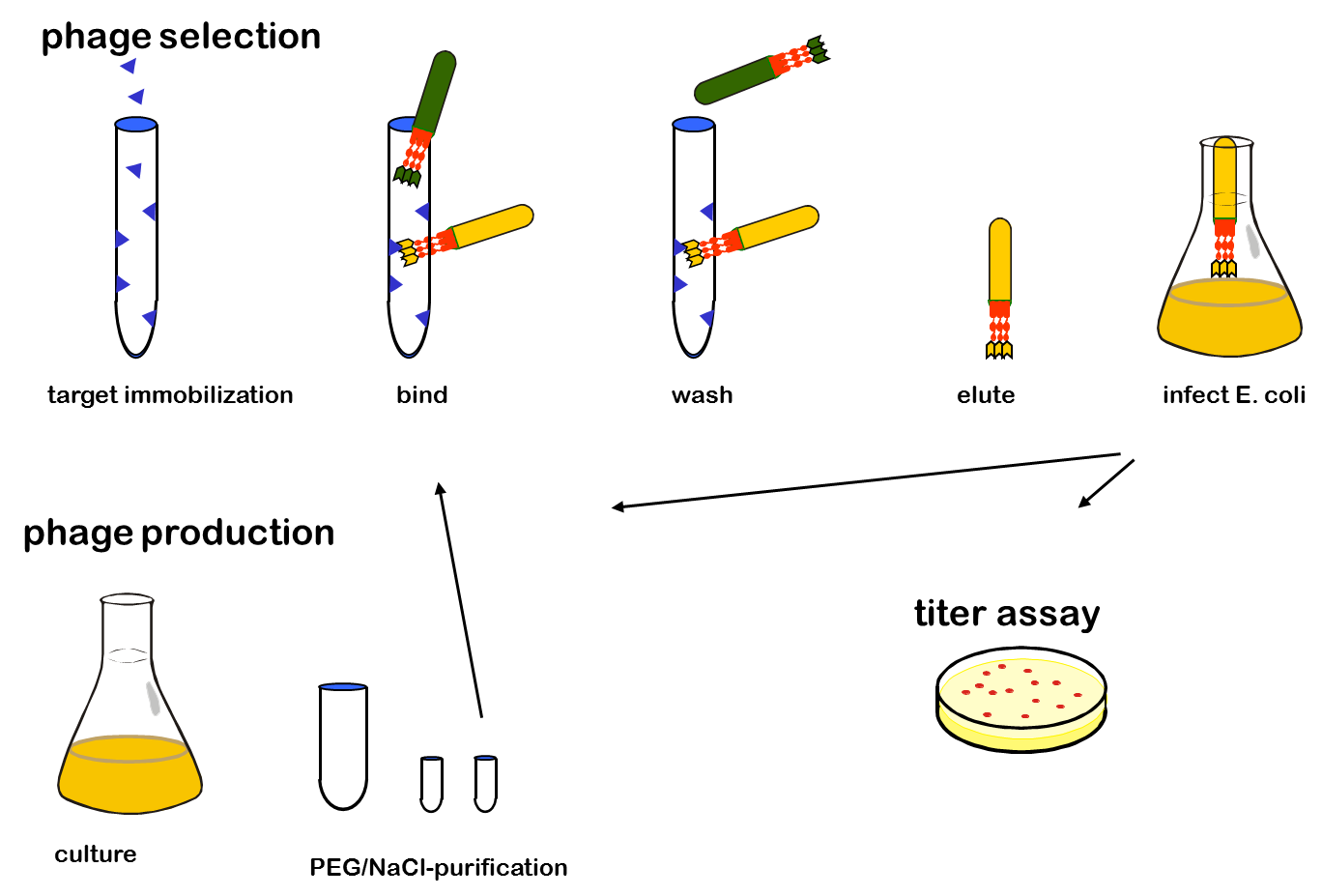
Phage display is a powerful tool for selecting peptides or proteins that bind and regulate the function of target proteins. It is defined as a system in which the protein and its encoding gene are physically linked. The coupling of peptides to phages and the subsequent selection of binders was first described by George P. Smith in 1985. The bacterial viruses principally used are members of the single-stranded filamentous phage family M13. After attachment to the bacterial F-pilus and entering the cells this phages stripe of their protein coat and convert their DNA in the double stranded form. Than replication, expression and assembly of phage particles occurs. About 200 phages can be produced per cell and generation. The M13 phage is a non-lytic phage, that means that they can be released from bacterial cells without causing cell death.
In M13 phage display genes from a DNA-library are usually fused to the gene III coat protein which occurs in five. The gene III protein is responsible for phage infection and releasing phage particles after production (Holliger and Riechmann, 1997; Stengele et al., 1990). Along with insertion of the gene of interest into the phage genome (McCafferty, 1990) phagemid vectors are established (Mead, 1988). Phagemids contain phage and bacterial origins of replication, the gene encoding the coat protein for fusion and a marker gene. To produce functional phages, co-infection with helper phages is required. These plasmids contain all genes used for producing phage particles. Its genome carries a mutation which leads to more efficient packaging of the phage genome of interest in relation to the helper plasmid genome.
One advantage of phage display is the rapid identification and concentration of target binding proteins through a procedure called biopanning. Bound phages are enriched by consecutive cycles of incubation, washing, amplification and selection. Therefor the target molecules are immobilized on ELISA plates or immune tubes. After an appropriate number of panning rounds, the inserts from the enriched phage can be sequenced to identify the interacting proteins.
 "
"