Team:HokkaidoU Japan/Project/T3SS
From 2011.igem.org
| Line 18: | Line 18: | ||
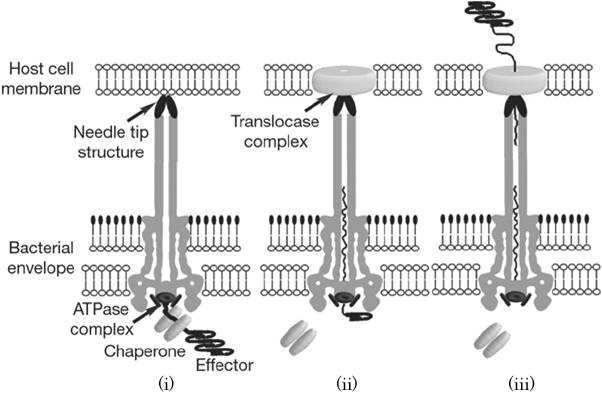
specific chaperone, and forms effector-chaperone complex. This complex is recognized by the T3-secretion-associated ATPase locatedat the base of the needle complex (i). Then, the ATPase stripes the chaperone from the complex and unfolds the effector protein to pass through the channel of the needle complex (ii). Finally, the translocated effector is refolded within the Target cell to carry out their function (iii)<sup>[2]</sup>. | specific chaperone, and forms effector-chaperone complex. This complex is recognized by the T3-secretion-associated ATPase locatedat the base of the needle complex (i). Then, the ATPase stripes the chaperone from the complex and unfolds the effector protein to pass through the channel of the needle complex (ii). Finally, the translocated effector is refolded within the Target cell to carry out their function (iii)<sup>[2]</sup>. | ||
| - | + | <div style="clear:both;"></div> | |
| + | =Our achievements on iGEM 2010= | ||
| + | [[Image:HokkaidoU_T3SS_Fig3.jpg|300px|thumb|left|Fig.3<br>A model of GFP injection assay]] | ||
==Construction of ''E. coli'' base GFP injector== | ==Construction of ''E. coli'' base GFP injector== | ||
| - | [[ | + | We made a T3SS test construct on pSB1T3 vector, and transformed it to the ''E. coli'' (K-12) carrying BAC vector "pBAC-SPI-2" that encodes a part of Salmonella enterica serovar Typhimurium LT2 genome, which would express T3S apparatus. |
| + | |||
| + | ==Secretion signal== | ||
| + | N-terminus 191 a.a. polypeptide functions as a T3 secretion signal domain in Salmonella<sup>[[#References|[6]]]</sup>. | ||
| + | |||
| + | ==NLS (Nuclear Localization Signal)== | ||
| + | In addition to the signal peptide, we located triple NLS (Nuclear Localization Signal) repeats between the T3 secretion signal and GFP so that the injected GFP would localize inside of the nucleus in the target cell. | ||
| + | |||
| + | ==RFP reporter== | ||
| + | The reporter gene constitutively produces RFP as an internal control that cannot be injected into target cells. This RFP would also play a role to distinguish GFP located in eukaryotic cells from GFP remaining in bacterial cells by indicating it as yellow color by fluorescence Microscopy. | ||
| - | + | ==Arabinose promoter== | |
| + | This [T3signal-NLS-GFP] fusion gene is under control of inducible arabinose promoter. If arabinose is added to the culture medium, ''E. coli'' produces both GFP and RFP so that the bacteria become yellow. However, if arabinose is not added, only RFP is produced so that the bacteria become red. | ||
Revision as of 11:45, 21 September 2011
Contents |
T3SS
Introduction
Type III Secretion System (T3SS) is a system of pathogenic gram-negative bacterium such as Salmonella, Yersinia and EPEC(entero pathogenic E. coli). Using this system bacteria can inject whole protein molecules through a syringe like organelle named Type 3 Secretion Apparatus (Fig. 1). The target of this system is a eukaryotic cell. Naturally it is used to inject Virulence effector proteins.
Structure of Type III secretion apparatus
Type III secretion apparatus have a syringe like structure. It can be visible under electron microscope [2]. Its length is about 80 nm and the diameter of its needle channel is about 2 nm [3]. The length is about 1/10 and the diameter is about 1/400 of an E. coli cell’s minor axis. Thus this is the smallest injector in the world (Fig.1 a~c).
How does it function?
When the needle tip attaches to the Target cell membrane, a translocator complex is assembled on the Target cell membrane. On the other hand, an effector protein which has a unique T3SS secretion signal domain on its N-terminal is recognized by the specific chaperone, and forms effector-chaperone complex. This complex is recognized by the T3-secretion-associated ATPase locatedat the base of the needle complex (i). Then, the ATPase stripes the chaperone from the complex and unfolds the effector protein to pass through the channel of the needle complex (ii). Finally, the translocated effector is refolded within the Target cell to carry out their function (iii)[2].
Our achievements on iGEM 2010
Construction of E. coli base GFP injector
We made a T3SS test construct on pSB1T3 vector, and transformed it to the E. coli (K-12) carrying BAC vector "pBAC-SPI-2" that encodes a part of Salmonella enterica serovar Typhimurium LT2 genome, which would express T3S apparatus.
Secretion signal
N-terminus 191 a.a. polypeptide functions as a T3 secretion signal domain in Salmonella[6].
NLS (Nuclear Localization Signal)
In addition to the signal peptide, we located triple NLS (Nuclear Localization Signal) repeats between the T3 secretion signal and GFP so that the injected GFP would localize inside of the nucleus in the target cell.
RFP reporter
The reporter gene constitutively produces RFP as an internal control that cannot be injected into target cells. This RFP would also play a role to distinguish GFP located in eukaryotic cells from GFP remaining in bacterial cells by indicating it as yellow color by fluorescence Microscopy.
Arabinose promoter
This [T3signal-NLS-GFP] fusion gene is under control of inducible arabinose promoter. If arabinose is added to the culture medium, E. coli produces both GFP and RFP so that the bacteria become yellow. However, if arabinose is not added, only RFP is produced so that the bacteria become red.
Bacteria and cell culture condition
To perform the injection assay, we used LB medium (1.0% Bacto-Tryptone, 0.5% Bacto-yeast extract, 1.0% NaCl) and magnesium minimal medium (MgM) [11] containing 170 mM MES-NaOH buffer(pH=5.0 or 7.2), 7.5 mM (NH4)2SO4, 5 mM KCl, 1 mM KH2PO4, 8 uM MgCl2, 38 mM glycerol and 0.1% casamino acids. We named these medium as MgM5(pH=5.0) and MgM7(pH=7.2). Also we used the acidic cell culture medium RPMI-10% FCS + HCl (pH 5.0) [RPMI5] and normal RPMI-10% FCS [RPMI7]. Bacteria were cultured at 37C with aeration and RK13 cells were cultured at 37C in 5.0% CO2. Appropriate antibiotics were added according to the resistance marker on each plasmid (25 ug/mL of chloramphenicol and 15 ug/mL of tetracycline). To induce GFP fusion protein L-arabinose was added to the medium at each step (final concentration = 0.4% ).
Methods of Injection Assay
Model for control of effector translocation by the SPI-2 T3SS. (i) Following uptake into host cells, acidification of the vacuole lumen induces assembly of 9 the secretion apparatus. (ii) Membrane-associated SsaL/SsaM/SsaB (Fig.3) regulatory complex (in purple, black and blue, respectively) prevents premature secretion of effectors (in brown). Translocon proteins (in green), connected to the T3SS apparatus, form a pore in the vacuolar membrane. (iii) The pore enables a component(s) of the T3SS to sense the elevated pH of the host cell cytosol, and a signal is transduced to the SsaL/SsaM/SsaB complex, which dissociates. (iv) Relief of effector secretion suppression enables their translocation [10].
As mentioned before the T3SS encoded in SPI-2 naturally function inside of the phagosome of the target cell [8]. So, it requires acidic pH to be assembled functionally [7]. However, if the T3 apparatus is assembled successfully under low pH(pH=5.0) condition, only the translocator proteins are secreted through T3SS but effector proteins are not. And it was reported in 2010 that the translocator complex assembles a pore on the phagosome membrane of the host cell enabling the T3SS to sense the neutral pH condition of the cytosol, and this pH elevation switches the function of the T3SS to start secretion of effectors (Fig.6) [10]. In addition we found that initial growth in MgM7 before the growth in MgM5 improve the production of GFP fusion protein in E. coli (data not shown). So, 10 hrs before exposure we transferred E. coli [SPI-2+GFP-T3signal+RFP] overnight culture from LB + arabinose to MgM7 + arabinose and grow for 4 hrs to charge sufficient amount of GFP fusion protein. 5.5 hrs before exposure, bacteria were transferred to MgM5 + arabinose and grow for 4 hrs to assemble T3 secretion apparatus. 1 hr before exposure, bacteria were washed with RPMI5 three times to remove toxin secreted from E. coli. Then it was resuspended diluted with RPMI5 + arabinmose (final ΔOD = 0.06 at 600 nm). On the other hand RK13 cells were seeded on 6-well plate (2x 10<sup5</sup> cells/well) in antibiotics free RPMI7 at 20 hrs before exposure to the E. coli. When the preparation is completed cell culture medium was replaced with 1 mL of the E. coli suspension (ΔOD = 0.06 at 600 nm) and incubate at 37C in 5.0% CO2 to mimic the environment inside of the phagosome. At the same time samples of arabinose(-) E. coli[SPI-2+GFP-T3signal+RFP] and E. coli[SPI-2 only] were prepared for the control condition. </div>
References
- Chen LM, Briones G, Donis RO, Galán JE. 2006. Optimization of the delivery of heterologous proteins by the Salmonella enterica serovar Typhimurium type III secretion system for vaccine development. Infect Immun. Vol.74:5826-5833. [http://www.ncbi.nlm.nih.gov/pubmed/16988261 PubMed]
- Galán JE, Wolf-Watz H. 2006. Protein delivery into eukaryotic cells by type III secretion machines. Nature. Vol.444:567-573. Review. [http://www.ncbi.nlm.nih.gov/pubmed/17136086 PubMed]
- Ghosh P. 2004. Process of protein transport by the type III secretion system. Microbiol Mol Biol Rev. Vol.68:771-795. Review.[http://www.ncbi.nlm.nih.gov/pubmed/15590783 PubMed]
- Hansen-Wester I, Chakravortty D, Hensel M. 2004. Functional transfer of Salmonella pathogenicity island 2 to Salmonella bongori and Escherichia coli. Infect Immun. Vol.72:2879-2888. [http://www.ncbi.nlm.nih.gov/pubmed/15102800 PubMed]
- Jacobi CA, Roggenkamp A, Rakin A, Zumbihl R, Leitritz L, Heesemann J. 1998. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol Microbiol. Vol.30:865-882. [http://www.ncbi.nlm.nih.gov/pubmed/10094634 PubMed]
- Miao EA, Miller SI. 2000. A conserved amino acid sequence directing intracellular type III secretion by Salmonella typhimurium. Proc Natl Acad Sci U S A. Vol.97:7539-7544. [http://www.ncbi.nlm.nih.gov/pubmed/10861017 PubMed]
- Rappl C, Deiwick J, Hensel M. 2003. Acidic pH is required for the functional assembly of the type III secretion system encoded by Salmonella pathogenicity island 2. FEMS Microbiol Lett. Vol.226:363-372. [http://www.ncbi.nlm.nih.gov/pubmed/14553934 PubMed]
- Waterman SR, Holden DW. 2003. Functions and effectors of the Salmonella pathogenicity island 2 type III secretion system. Cell Microbiol. Vol.5:501-511. Review. [http://www.ncbi.nlm.nih.gov/pubmed/12864810 PubMed]
- Wilson JW, Coleman C, Nickerson CA. 2007. Cloning and transfer of the Salmonella pathogenicity island 2 type III secretion system for studies of a range of gram-negative genera. Appl Environ Microbiol. Vol.73:5911-5918. [http://www.ncbi.nlm.nih.gov/pubmed/17675443 PubMed]
- Yu XJ, McGourty K, Liu M, Unsworth KE, Holden DW. 2010. pH sensing by intracellular Salmonella induces effector translocation. Science. Vol.328:1040-1043. [http://www.ncbi.nlm.nih.gov/pubmed/20395475 PubMed]
- Yu XJ, Liu M, Holden DW. 2004. SsaM and SpiC interact and regulate secretion of Salmonella pathogenicity island 2 type III secretion system effectors and translocators. Mol Microbiol. Vol.54:604-619. [http://www.ncbi.nlm.nih.gov/pubmed/15491354 PubMed]
 "
"