Team:Debrecen Hungary/Project
From 2011.igem.org
(→Lipid Sensors: Nuclear Hormone Receptors) |
(→Lipid Sensors: Nuclear Hormone Receptors) |
||
| Line 156: | Line 156: | ||
[[File:CheesepparNHR8.png|center|600px|thumb|Activation of PPARg to Bresse bleu cheese extract in different cell lines.]] | [[File:CheesepparNHR8.png|center|600px|thumb|Activation of PPARg to Bresse bleu cheese extract in different cell lines.]] | ||
| - | [[File:NHR8cheese.png|center| | + | [[File:NHR8cheese.png|center|400px|thumb|Activation of NHR8 to Blesse bleu cheese extract.]] |
=== Effector: Gene expression regulated by zinc fingers === | === Effector: Gene expression regulated by zinc fingers === | ||
Revision as of 07:29, 21 September 2011
Abstract
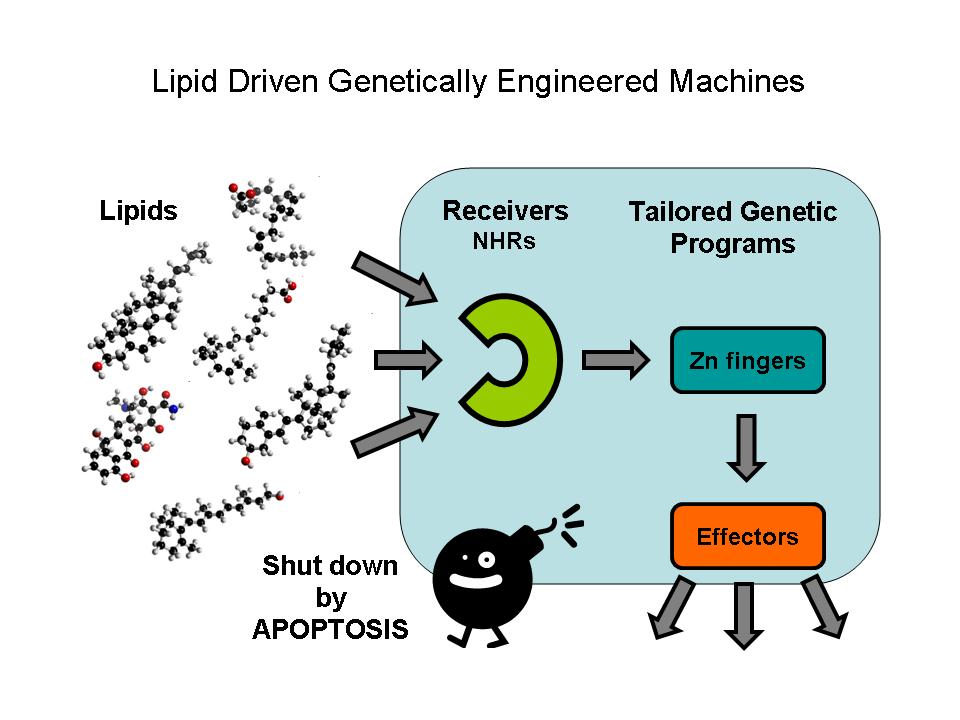
Nuclear hormone receptors (NHRs) are ligand activated transcription factors. They are able to regulate the expression of their target genes by direct DNA-binding, in a ligand-dependent manner. NHRs bear high homology to each other and are modular into distinct regions: N-terminal regulatory region, DNA-binding domain, a Hinge region, Ligand binding domain (LBD) and a C-terminal region. Some Nematode NHRs can be activated by extracted oil contamination of the soil so they can use as possible oil sensors in the future. Zinc finger motifs are the tools of NHRs to bind DNA and regulate gene expression directly. These tiny elements can be tested as direct gene regulators. Controlled gene induction can also lead to programmed cell death which is a less harmful tool in order to quit non-functionable cells.
Project description
Lipids constitute a broad group of naturally occurring molecules which include fats, waxes, sterols, fat-soluble vitamins (such as vitamins A, D, E and K), monoglycerides, diglycerides, phospholipids, and others. The main biological functions of lipids include energy storage, as structural components of cell membranes, and as important signaling molecules.
Although there are certain types of lipids their specific investigation is very difficult. Sophisticated chromatographies, mass spectrometry, nuclear magnetic resonance (NMR) spectroscopy, fluorescence spectroscopy or dual polarisation interferometry are used in certain lipidomics centers. Another possibility is to investigate their specific regulatory effects. The receivers are nuclear hormone receptors, which is a conserved family of proteins responsible for sensing lipids; they may be viewed as lipid activated transcription factors.
Ligand binding domains respond only to predefined lipids and selectively activate predetermined gene expression. The tool for regulating directly the gene expression of the DNA binding domains of nuclear hormone receptors are tiny zinc finger motifs. These elements bind straight to the DNA and can act as direct gene expression regulators so they can operate as specific effectors.
A machine has to be switched off after using it. In our case the power button means that the genetically modified cells have to be removed without any harm caused in the system. A perfect mechanism for that is inducing Programmed Cell Death in a controlled way.
The parts designed by Team: Debrecen-Hungary 2010 allows the ultra sensitive detection of xenobiotic activators in the environment (e.g. oil derived traces in water and soil), the assessment of lipid soluble vitamins, steroids, xenobiotics etc. in food and the potential of inducing specific genetic programs in transfected cells or model organisms with a possibility to destroy these modified cells by an independent induced apoptosis effector system.
Our team decided to work with the parts modelled by last year's team in order to further characterize and biuld new composite parts and expression vectors from them which led to compose a lipid driven genetically engineered machine.
Basics
Synthetic Biology
Synthetic biology is a relatively new area of biological research; similar to many other new scientific fields it has many definitions. The best way to express the meaning of synthetic biology is to understand the desired end-result: engineering of complex biological systems. These systems are best thought of as analogues of everyday machinery: cogwheels, levers, timers, button and buzzers (in this case a clock), only in the case of biological systems (molecular ones) cells, DNA, proteins, lipids, sugars and RNA are the “parts” of the system.
Similar to mechanical engineering (or every other engineering branch) there is a need for standards (consensus way of doing things), abstraction (simple and unified way of thinking about the parts of a system), and modularity (how these parts interact to become the device, or several devices into a system). Thus a good definition for synthetic biology could be engineering of molecular (for the time being) biological systems according to preset standard parts. The international genetically engineered machines and associated parts registry are, to date, one of the largest registries for standard parts in use for synthetic biology. Free information can be found in the registry regarding parts, devices and modules all inputted by various teams worldwide.
Eukaryotic synthetic biology is still in its infancy. The large kingdom of metazoans includes all multicellular eukaryotes such as mammalians, arthropods and nematodes. No standard chassis (framework) exists for the animal kingdom which makes them far less popular then the famous E.coli. The protein modules derived from metazoans (like Drosophila or C. elegans) are functional in yeasts also. Very few iGEM teams (or even labs outside iGEM) have chosen to toggle the animal chassis (two notable examples are team Heidelberg 2009 and team Slovenia 2006). The amount of available compatible parts is limited, which severely restricts the options of creating complex biological devices. Nearly no imagination is required for designing tools, since their analogues already exist in the bacterial chassis. The possible use of such systems is unlimited. Field’s such as of environment, medicine, energy and research all gain to profit from the development of animal synthetic biology.
Systems requiring gene expression input in eukaryotic synthetic biology systems require a way to standardize gene expression, a complicated task. The way from gene to protein contains many steps of possible error: transcription factor binding, promoter strength, recruitment of auxiliary proteins, nuclear RNA synthesis and many more steps finally leading to translation, folding, cleaving and delivery (but hey, you have to start somewhere).
Our team was interested at designing eukaryotic synthetic biology tools related to PoPs. PoPs (polymerase per second), the flying Dutchman of synthetic biology, is a number which represents the rate (base pair per second) at which RNA polymerase crosses past a given DNA position. Currently, no in vivo technique for measuring PoPS directly exists; it can be estimated indirectly by measuring other parameters (eg protein expression or enzyme activity). Nevertheless it is still a useful abstraction for thinking about transcription-based logic devices and it allows the engineer to define devices. Our aim, was not only to infer PoPs but to devise a way to titrate it remotely.
Nuclear Hormone Receptors
Nuclear receptors are ligand activated transcription factors. As such, they are able to regulate the expression of their target genes by direct DNA-binding, in a ligand-dependent manner. They play a central role in endocrine signaling, regulation of embryonic and adult development and differentiation. Many nuclear receptors, are among the primary targets of drug discovery because of their diverse biological actions. Nuclear receptors bear high homology to each other and are modular into distinct domains: N-terminal regulatory domain, DNA-binding domain, a Hinge region, Ligand binding domain (LBD) and a C-terminal domain.
Ligand binding domain (LBD) is a well conserved domain amongst various nuclear receptors whose structure usually referred to as an alpha helical sandwich fold. The LBD shows some diversity among nuclear receptors as it is a site for receptor-specific events. It possesses transactivation ability and contains a ligand-binding pocket as well as the main interaction surfaces for other proteins.
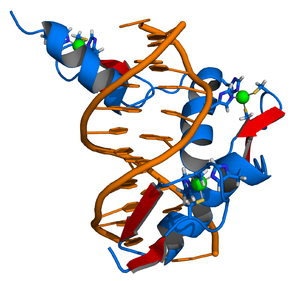
The DNA-binding domain (DBD) contains two Zinc-finger motifs and is linked to the LBD by a highly flexible hinge region. This segment of the nuclear receptors holds the ability to recognize and bind to preferred or specific DNA-motifs, the response elements.
A partially unexpected and amazing feature of the nuclear receptors is that the above mentioned domains can be swapped. It was Vincent Giguere and his colleagues, who first devised a technique called cotransfection assay and developed it to the domain-swap technique.
Zinc Fingers
Zinc fingers are small protein structural motifs binding one or more zinc ions to help stabilize their folds. Zinc finger proteins often bind to DNA and RNA because their shape allows close interaction of the domain with the nucleotides of DNA and RNA, and they can also bind to proteins and small molecules. Their nucleotide binding properties allow them to function in regulating gene expression and in virus assembly.
Zinc fingers coordinate zinc ions with a combination of cysteine and histidine residues. They can be classified by the type and order of these zinc coordinating residues (e.g., Cys2His2, Cys4, and Cys6). A more systematic method classifies them into different "fold groups" based on the overall shape of the protein backbone in the folded domain. The most common "fold groups" of zinc fingers are the Cys2His2-like (the "classic zinc finger"), treble clef, and zinc ribbon. The DNA binding domain of nuclear hormone receptors is a highly conserved domain containing two zinc fingers that binds to specific sequences of DNA called hormone response elements (HRE).
Caenorhabditis elegans
Caenorhabditis elegans is a free-living, transparent nematode (roundworm), about 1 mm in length, which lives in temperate soil environments C. elegans is intensively studied as a model organism in biology for a variety of reasons. The developmental fate of every single somatic cell (959 in the adult hermaphrodite; 1031 in the adult male) has been mapped out. The C.elegans genome harbors 284 nuclear receptors (a striking figure), which have been shown to control traits such as sex determination, larva development, life span, neuronal growth and identity and much more. As far as nuclear receptors go, they are a gold mine.
Apoptosis
Apoptosis, the process of programmed cell death (PCD), is an elaborate cellular homeostasis mechanism that ensures correct development and function of multicellular organisms. Biochemical events lead to characteristic changes (morphology) and the death of cells.
It has a crucial role in maintaining tissue homeostasis as provides the appropriate rate of cellular turnover. Potentially dangerous, poorly differentiated or excess cells are removed by apoptosis without local inflammation from leakage of cell contents. Dying by apoptosis in many cases has primary importance: lens cells, which lost their nucleus, need the vision, the corneocytes without nucleus and organelles provide mechanical protection on the outer surface of the skin.
Apoptosis might occur in pathological conditions, mainly in secondary, as a part of the defensive mechanism, helping to eliminate the viral infected or malignant cells. The abnormality of the central molecules and genes of apoptosis may play a role in the development of certain diseases: degenerative disease, autoimmune disease, tumor formation and malformations.
Many pathways and signals lead to apoptosis, but there is only one mechanism that actually causes the death of a cell. After a cell receives stimulus, it undergoes organized degradation of cellular organelles by activated proteolytic caspases. A cell undergoing apoptosis shows characteristic morphology.
Luciferase
Luciferase is an enzyme class able to produce bioluminescence by oxidizing the substrate luciferin. "Firefly luciferase" as a laboratory reagent usually refers to P. pyralis luciferase. Its emission can be measured photometrically and hence used to deduce the protein enzyme concentration through standardized methods.
Cos-1 cells
Cos-1 cells (acronym for CV-1 simian origin, SV-40 viral positive) is cell line derived from the African green monkey kidney cells. It is also often used to transfect cells in tissue culture conditions to produce recombinant proteins for molecular biology, biochemistry, and cell biology experiments. Two forms of COS cell lines commonly used are COS-1 and COS-7. The cell line was obtained by immortalizing the original CV-1 cells with SV-40 virus genome. This allows the production of large T antigen but has a defect in genomic replication.
The Experiments
Cotransfection Assay, Two Hybrid Assay
Cotransfection assay requires two types plasmids to be cotransfected into a cell; an expression vector, coding a functional NR and and a reporter-plasmid, harboring an inducible promoter that regulates a reporter-gene the expression level of which can easily be detected and quantified. By this method the NR-activity and ligand-potency can be studied. As a result of the unique domain-structure of NRs, the domain-swap technique was developed, using chimeric proteins; the DBD of a known receptor is fused to any other receptor. This made ligand screening possible.
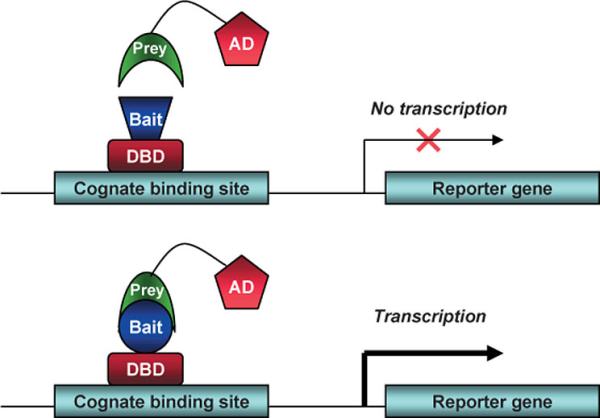
The two-hybrid assay is a technique in molecular biology which can be used to investigate protein-protein interactions. The interaction of a DNA-binding bait-protein and the prey-protein with a ligand binding and activating ability is shown by the activated reporter.
Multiple applications of these methods have appeared. It is possible to investigate:
- the response elements by changing the sequence that is recognized by the DBD
- potential dimer partners and co-regulators by creating specific bait-prey sets
- potential ligands and activating factors
Our main concept was to introduce the iGEM-method into this well established area of research. We showed that these systems (they were published before) can be reproduced with the use of our pre-designed Parts. We could also present that it is possible to take the next step: to investigate new samples as well, i.e. to test the effects of different biological samples on NR-action by our Kits.
This image shows the basic concept of the two-hybrid system: the DNA-binding bait-protein (Gal-fused, in our case), and the activator-domain fused to the prey-protein (mostly VP-fusion, in our case) and the reporter gene with the specific binding site. Yeast Gal4 is a common DBD used for this techniques purpose. Commonly used reporter genes include the product of the LacZ gene (Beta galactosidase) and Luciferase.
Results
Cloning
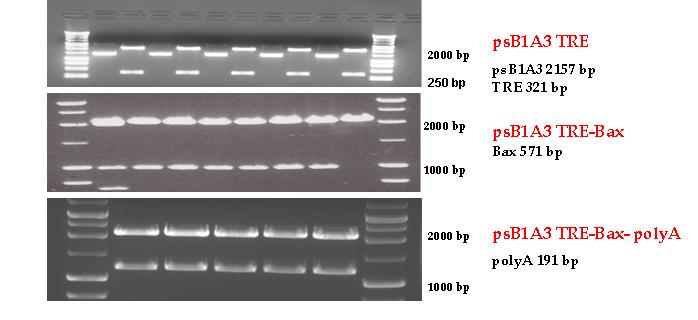
Firstly, we had to generate expression vectors that can be used in mammalian expression system. The cloning work of Team Debrecen-Hungary 2011 resulted 3 composite parts and one fully synthetic biology comaptible expression vector from pSB1A3. All of them have been tested by restriction enzyme analysis and by sequencing. Furthermore we have tested two Zn-fingers and fusion vectors by luciferase assay.
NHR64-GAL4 fusion
NHR64 LBD of C. elegans has been inserted after Gal4 in pSB1C3, resulting an insert with the lenght of 1008 bp. After that NHR64-GAL4 was transferred into pCDNA3.1 to express it in certain cell lines.
CD38-EcR LBD-pSB1C3/ ADRP-EcR LBD-pSB1C3 fusions
The CD38/ADRP (Modelled by Team: Debrecen-Hungary 2010) are Zinc-finger proteins that can bind to the DNA, and CD38/ADRP (530bp) were fused with EcR-LBD. The EcR (768 bp) is a nuclear receptor that is the ortolog of mammalian RXR and contains ligand binding domain. After the fusion, the insert length was 1298bp. Then CD38/ADRP-EcR LBD were transferred into pCDNA3.1.
BAX expressing vector from pSB1A3
Three basic parts from pSB1C3 have been ligated into pSB1A3 consecutively by using chloramphenicol and ampicillin assembly in order to create an expression vector from the ampicillin resistance carrying backbone. The first step was to insert 321 bp long TRE-CMV (Tetracyclin Response Element- Minimal Cytomegalovirus promoter) into pSB1A3 which has the length of 2157 bp. Then BAX (573 bp) were inserted after the TRE- CMV, before PolyA tail (191 bp) was ligated in the end. As the negative control of the expression vector built up by Team: Debrecen-Hungary 2010 from standard biobrick parts TRE- CMV and PolyA tail were ligated together into pSB1C3 and pSB1A3.
Lipid Sensors: Nuclear Hormone Receptors
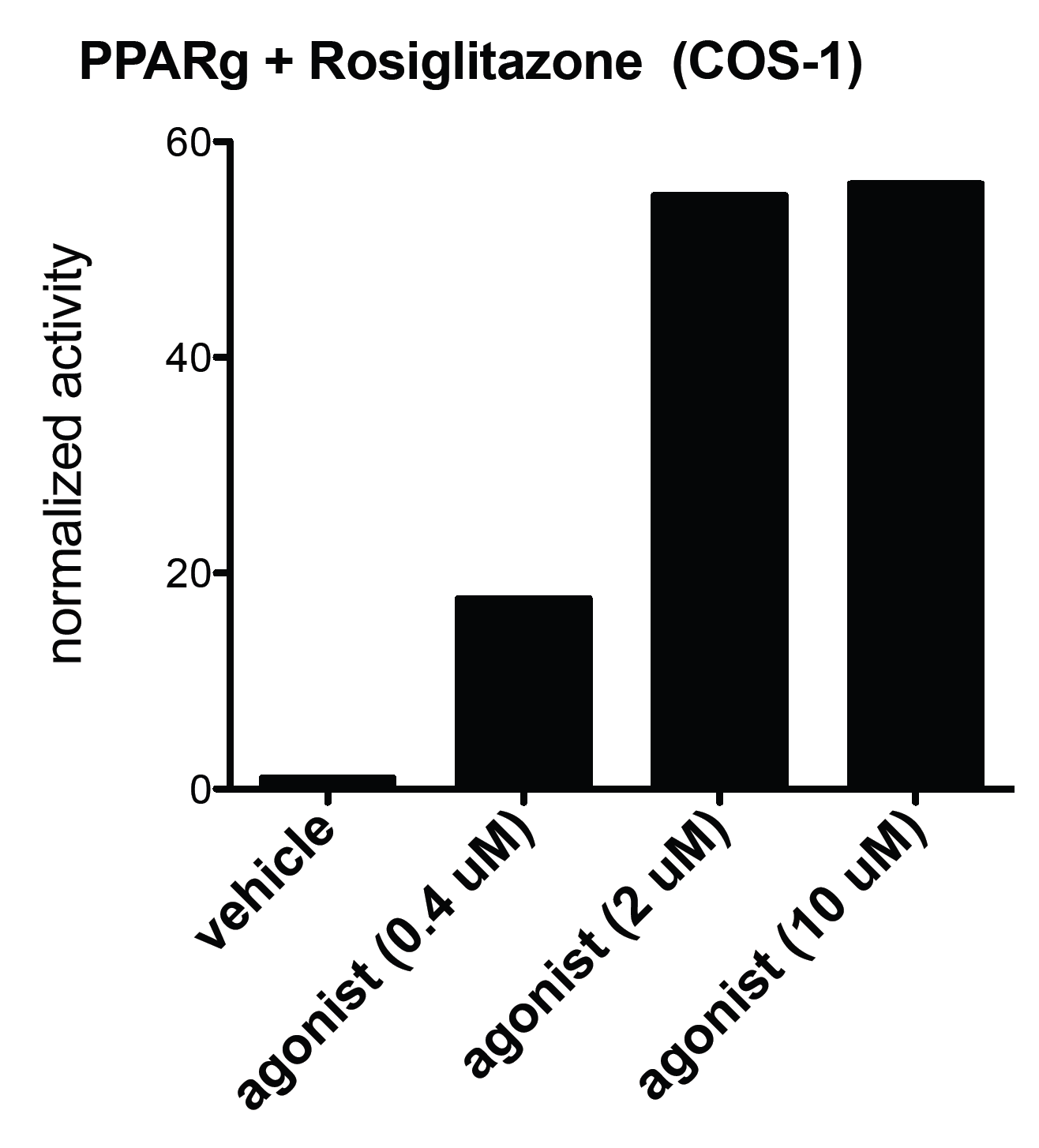
To test our system, we transfected the Gal-PPARg plasmid into COS-1 and 293T cell lines and applied an agonist treatment (Rosiglitazone,BRL). The expressed Gal-PPARg protein was able to bind to the UAS-sequence of the co-transfected luciferase reporter (MH100-TK-luciferase) and activate it in a dose dependent manner. This agonist is a higly potent activator of the PPARg, which explains the high level of luciferase activity.
As the set up system was ready to test PPARg-ligands, we next tested oil-extracts (extracted from oil sand samples). Suprisingly, the treatment with the oil-extract caused dose dependent activation of PPARg. The level of activation was clearly lower than the one we could detect with the specific agonist, but the dose dependence gave us a hint that the oil extract can activate the PPARg nuclear receptor.
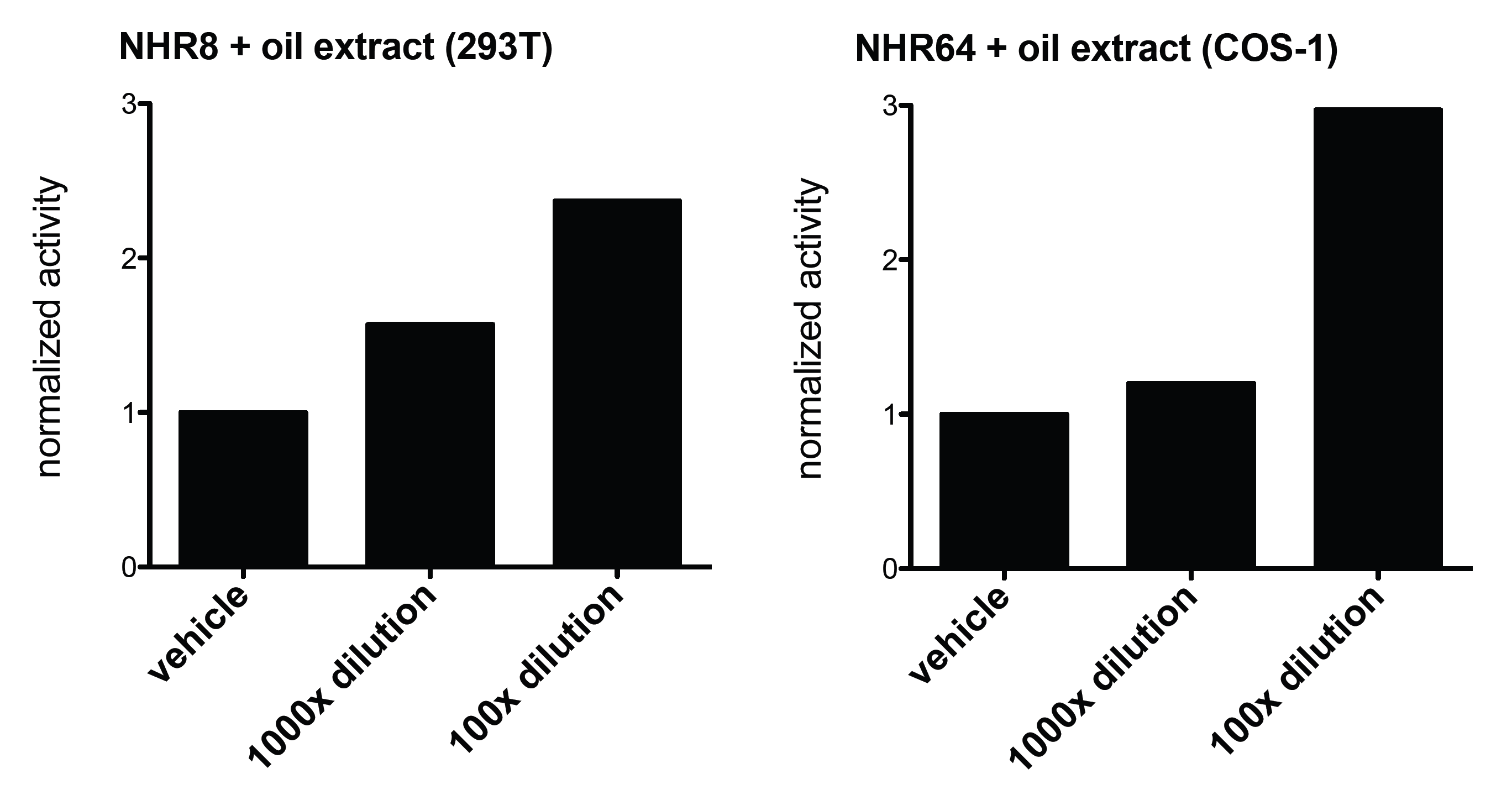
Our idea was to apply the nuclear receptors as parts of a system built up from sensors and effectors. As lipids are relatively hard to detect and investigate and some classes of the nuclear receptors are specialized to lipid sensing in living organisms, we gained a search for potential oil sensors. This is how we came accross with the C. elegans worm and its nuclear receptors (NHR). Due to its living habitat the nuclear receptors of this worm seemed to be good candidates for new oil-sensors. We created plasmids (using iGEM-standards) by the fusion of a Gal UAS-binding domain and a NHR (Gal-NHR8, Gal-NHR64). No agonist ligands are known for these receptors, so far.
Next, we transfected the Gal-fused worm nuclear receptors (NHR8, NHR64) into mammalian cell lines along with the other parts of the luciferase reporter system, treated the cells with the oil extract and measured the luciferase activity. Latter was the result of and activated luciferase gene, that was activated by the Gal-NHRs bound to the UAS, which became active as a result of the oil-treatment. Constructs baring NHR8 or NHR64 showed dose dependent activation upon the addition of the oil extracts to the cells. These NHRs acted as potential oil-sensors on which even a mammalian-cell based detection system can be built.
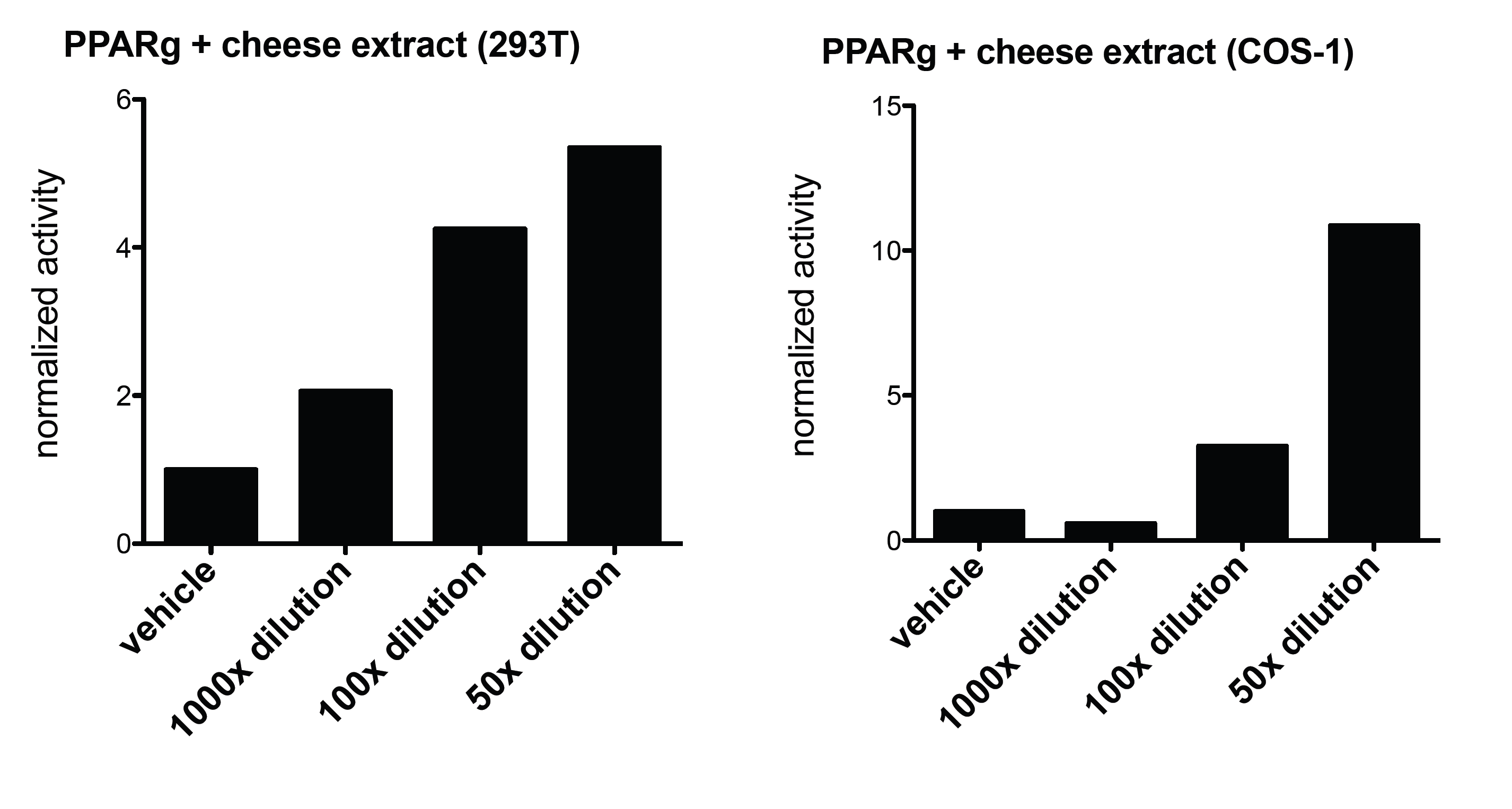
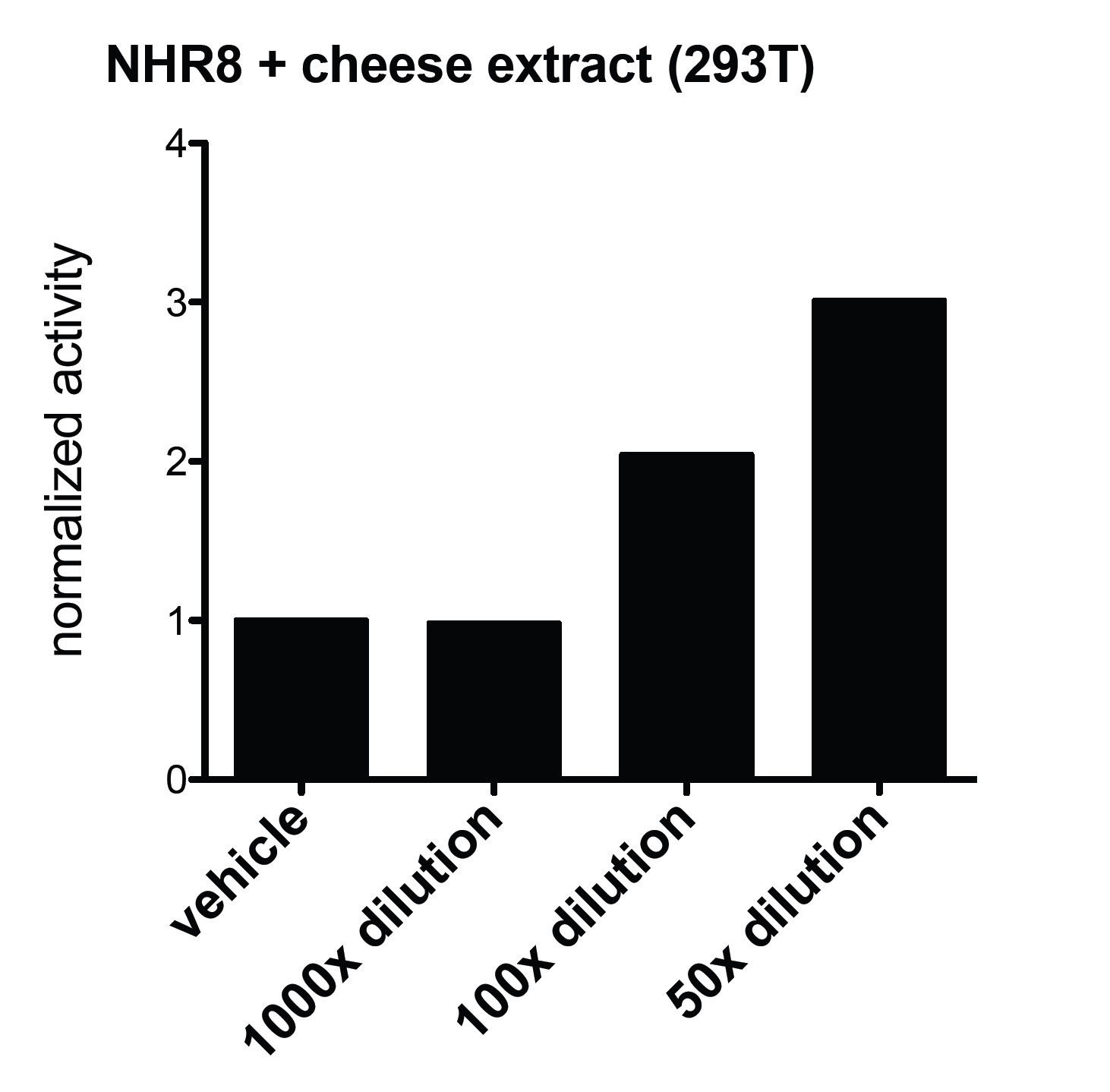
We were curious, wether other types of lipids could be detected by our system, so next we tested lipids not from the soil, but from food. Lipids were extracted from different types of cheese. We used above mentioned system to carry out similar experiments, but this time with cheese extracts. We could detect PPARg activity in each used cell line after the treatment with the Bresse bleu extract. The activity was dose dependent. This is not just the sign of a molecule of lidid nature in the extract, but a molecule that acts as a PPARg agonist. Even more suprisingly, this extract could activate the Gal-NHR8 system as well. It is important to mention though, that it was not necessarily the same compound of the cheese extract that showed these activities on different receptors.
Effector: Gene expression regulated by zinc fingers
One important feature of most of the nuclear receptors is that they are built up of well conserved protein motifs, domains. Still these motifs are the key to their specific action. The specificity of the ligand binding domain (LBD) was just shown earlier by testing different LBDs for sensing activating agents in oil and cheese extracts. The other domain that is responsible for selectivity is the DNA binding domain (DBD). In our previous experiments the DBD was replaced with a Gal-domain, that was necessary for the lucferase system when the LBD was being tested.
To show that even „imaginary” NRs can be built by the fusion of a potent LBD and a well designed DBD, we started to design a NR-specific DBD that contains a Zn-finger motif that is able to recognize a consensus retinoic acid receptor (RAR) response element (the region of an RAR target gene that the RAR is able to recognize and bind to). The construct is a newly designed Zn-finger motif. The design was based on the promoter region of CD38 gene (an RAR target gene). The Zn-finger sequence was then inserted into the pcDNA vector (ZN-FINGER). The DNA binding capacity of this protein was tested on a DR5-luciferase reporter (that carries a consensus RAR-binding sequence) by a competition experiment.
The DR5-sequence of the reporter could be recognized by the endogenous RAR of our cell lines, as it is shown by the base activity. If our ZN-FINGER was able to bind to the same sequence, a competition should occur between the endogenous RARs and our ZN-FINGERS. As the basal activity was reduced with increasing the amount of the transfected ZN-FINGER plasmids, we concluded that the designed protein can bind to the DR5 (Fif.8.). To show the effect on an enhanced level, we activated the system (by AM580 – RAR agonist ligand). By this, even larger the activity grew without the ZN-FINGERS. But again, by adding the ZN-FINGERS to the system, the level of activity was reduced (Fig.9). The numbers show the ratio between the ZN-FINGERS and the buffer plasmids:
1. ZN-FINGERS:buffer plasmids-1:8
2. ZN-FINGERS:buffer plasmids-1:2
3. Only ZN-FINGERS
By these experiment we have shown that based on the NR-system a great variety of sensor-effector system can be built. The different LBDs give the arsenal of sensing. From dietary components to different elements of the soil. The other side is the effector side that –via the different DBDs- can also be extremely variable. These sides are potentially switchable; different LBDs with different DBDs. Even at interspecific level.
Shut down: Apoptosis inducing (mechanism to switch off cells in a controlable way)
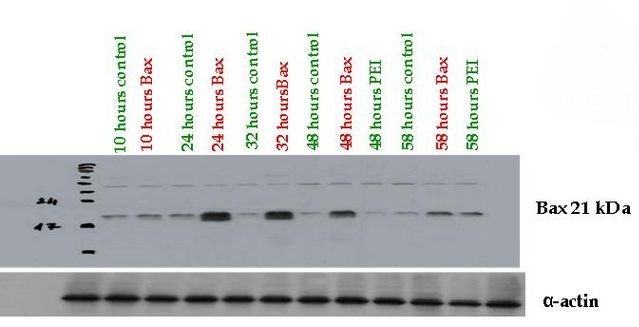
Fully Synthetic Biology compatible Bax proapoptotic protein expressing vector (BBa_K626000) was assessed from pSB1A3 which was transfected into COS-1, 293T and THP-1 cells. The functionality of the expression vector was proved by immunoblotting by using mouse primary antibody against human Bax (Santa-Cruz Biotechnology) where high protein expression could be detected 24-48 hours after transfection compared to the control cells.
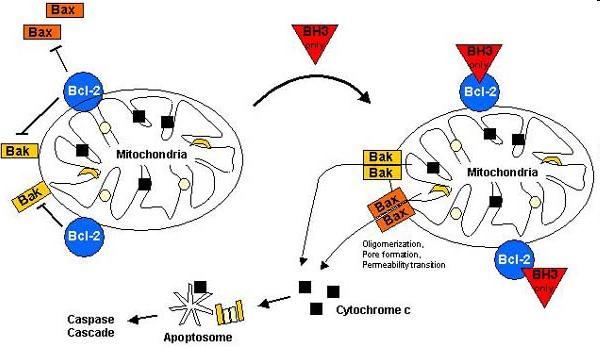
Bax (Bcl-2-associated X) protein is a pro-apoptotic member of the Bcl-2 protein family. It contains three BH (Bcl-2 homology) domains, can form homo- and heterodimers and plays essential role in the formation of MOMP (Mitochondrial outer membrane permeabilization) leading to apoptosis through cytochrome c release which is the most frequent type of PCD (Programmed cell death).
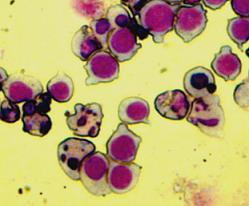
The Bax overexpression led to apoptosis in the transfected cells which was proved by DAPI (Sigma-Aldrich) staining. The nuclei of the apoptotic cells show fragmentation and bleb formation which are well-known markers of apoptosis. They also showed Annexin V positivity detected by flow cytometry.
Contributions to the project
All experiments were performed by our registered team members, and we all take responsibility for the contents of our wiki page and the parts uploaded by us to the Registry of Standard Biological Parts. For detailed contributions see the lists below.
Detailed contributions:
Finding out the project: was a collective decision.
Gene design: is made by our instructors and advisors, although they gave us the chance to learn how to design and order oligonucleotides.
Bacterial works, Cloning work: is mostly done by Undergraduate students, partly by Postgraduate masters students.
Transfections, cell works and ligand treatings: were carried out by the student members of the team.
Luciferase cotransfection assays: were done by the students with the help of one of our instructors.
Apoptosis project: were done by one of our instructors.
Arsenic project: done by one of our Undergraduates.
Wiki, Partsregistry and things related to registration: All of these works were carried out by the participation of almost all the members.
Contributions member by member:
Students (in alphabetical order)
Tímea Beregi: cloning, transfections, wiki writing.
Éva Bernadett Bényei: Our young high school student took part in cloning, bacterial and tissue culture works, and helped us to survive evenings in Lab by her stories and games during incubation times.
Beáta Boros-Oláh:She participated in all lab works including cloning, bacterial works, tissue culture works, Luciferase/cotransfection assays, and also worked on uploading matter to the wiki. Making photos of labwork.
Lilla Ozgyin: Cloning and tissue culture works, transfections, luciferase assays and writing matter to the Registry and coordinating the wiki mission. Protocol writing, arsenic biosensor testing, extraction of contaminated environmental samples.
Petra Sepsi: Our enthusiastic team member took part in every cloning, bacterial and tissue culture works, luciferase assays and she uploaded materials to the wiki. Making photos on labwork.
Ildikó Tándor: She helped us in every possible way, including cloning, bacterial and cell culture works, luciferase assays and wiki writing. Uploading materials to the Registry.
Advisors (in alphabetical order)
Péter Brázda: He supervised planning and evaluation of luciferase assays, participated in wet-lab education and the wiki-works. Mentored our high school student to get acquainted with the concept of synthetic biology. He also gave us useful advices on the implementation of processes in which the students had some difficulties.
Gergely Nagy: Gergely was a real advisor by giving us a dozen of tips on labwork, and taught us the scientific way of thinking. He gave us help in the implementation of results and directed us towards critical way of thinking with his fancy imagination as well.
Katalin Sándor: She was the mentor of our new students in the first few weeks, helping them to get to be in practice with basic laboratory methods, and she passed her experiences by giving presentations to us.
Instructors (in alphabetical order)
Bálint L. Bálint M.D. Ph.D.: He was our main coordinator by initiating the project, he made the team recruitment, fund raising, some of the PR and laboratory setup. He supervised the first lab experiments and planned some of the experiments later on, ordered parts and reagents, organized finances. He supervised wiki and partsregistry work as well. He dealed with fundraising.
Bence Dániel: He was a supervisor of the whole labwork, he gave us new directions when the lab work stucked by unexpected reasons. He shared his huge experience of nuclear receptors, trained our new students and helped us with the cotransfection assays. He also worked on the wiki project and uploaded materials to the registry. He performed the sequencing of our new parts.
Endre Károly Kristóf M.D.: Endre helped us in the apoptosis project (as he is experienced in examining apoptosis) and in the training of new students during the summer. He also gave help in writing an article about the team, writing the abstract, uploading files to the Registry and our team wiki. Participating in fund raising, in silico design, video and media project. Uploading materials of project description to the wiki and informations about the composite and some basic parts to the partsregistry.
Gábor Zahuczky Ph.D.: He supported the initiation of the project, fund raising, organization of finances and administration. Participated in the design of parts and wet lab education.
 "
"