Team:Potsdam Bioware/Labjournal/September part 1
From 2011.igem.org
KatharinaB (Talk | contribs) |
|||
| Line 1,883: | Line 1,883: | ||
** pSB1A3_YFP_Ara clone A | ** pSB1A3_YFP_Ara clone A | ||
| - | ** | + | ** pSB1A3_YFP_Lac clone B |
** pSB1K3_CFP_Ara clone A | ** pSB1K3_CFP_Ara clone A | ||
| - | ** | + | ** pSB1K3_CFP_Lac clone B |
| - | ** | + | ** pSB1K3_YFP_Lac clone C |
<b>Method:</b><br> | <b>Method:</b><br> | ||
| Line 2,935: | Line 2,935: | ||
* glycerol stocks of pARW089 and pARW071 | * glycerol stocks of pARW089 and pARW071 | ||
| - | * plates of pSB1C3 + mdnBC (from 2011-09-04), pUP089 A and B, pSB1A3_Ara_YFP, | + | * plates of pSB1C3 + mdnBC (from 2011-09-04), pUP089 A and B, pSB1A3_Ara_YFP, pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP, pSB1K3_Lac_YFP (from 2011-09-03) |
* LB medium | * LB medium | ||
| Line 4,241: | Line 4,241: | ||
* plate of pSB1A3_Ara_YFP (from 2011-09-03) | * plate of pSB1A3_Ara_YFP (from 2011-09-03) | ||
| - | * test cultures of | + | * test cultures of pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP (from 2011-09-06) |
* LB medium, Kan, Amp | * LB medium, Kan, Amp | ||
| Line 4,657: | Line 4,657: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">test pSB1A3_Ara_YFP and | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">test pSB1A3_Ara_YFP and pSB1A3_Lac_YFP (expression backbones) </h3> |
<b>Time:</b> 2011-9-7,12<br> | <b>Time:</b> 2011-9-7,12<br> | ||
| Line 4,665: | Line 4,665: | ||
<b>Method:</b> | <b>Method:</b> | ||
| - | * | + | * pSB1A3_Lac_YFP: |
**ODs at 16:45: 0.095 (1) and 0.082 (2, control) | **ODs at 16:45: 0.095 (1) and 0.082 (2, control) | ||
| Line 4,763: | Line 4,763: | ||
<b>Materials</b><br> | <b>Materials</b><br> | ||
| - | * plates of pSB1A3_Ara_YFP, | + | * plates of pSB1A3_Ara_YFP, pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP (from 2011-09-03) |
* plates of pUP089 A and B | * plates of pUP089 A and B | ||
| Line 5,067: | Line 5,067: | ||
** pSB1A3_Ara_YFP clone A (#1), resistance: amp | ** pSB1A3_Ara_YFP clone A (#1), resistance: amp | ||
| - | ** | + | ** pSB1A3_Lac_YFP clone B (#4), resistance: amp |
** pSB1K3_Ara_CFP clone A (#25), resistance: kan | ** pSB1K3_Ara_CFP clone A (#25), resistance: kan | ||
| - | ** | + | ** pSB1K3_Lac_CFP clone B (#21), resistance: kan |
| - | ** | + | ** pSB1K3_Lac_YFP clone C (#18), resistance: kan <br> |
* antibiotics from stock: | * antibiotics from stock: | ||
| Line 5,129: | Line 5,129: | ||
** pSB1A3_Ara_YFP clone A (#1), resistance: amp | ** pSB1A3_Ara_YFP clone A (#1), resistance: amp | ||
| - | ** | + | ** pSB1A3_Lac_YFP clone B (#4), resistance: amp |
** pSB1K3_Ara_CFP clone A (#25), resistance: kan | ** pSB1K3_Ara_CFP clone A (#25), resistance: kan | ||
| - | ** | + | ** pSB1K3_Lac_CFP clone B (#21), resistance: kan |
| - | ** | + | ** pSB1K3_Lac_YFP clone C (#18), resistance: kan |
* Ligation products stored in freezer, green box, mdn-BioBricks | * Ligation products stored in freezer, green box, mdn-BioBricks | ||
| Line 7,135: | Line 7,135: | ||
** pSB1A3_YFP_Ara | ** pSB1A3_YFP_Ara | ||
| - | ** | + | ** pSB1A3_YFP_Lac |
** pSB1K3_CFP_Ara | ** pSB1K3_CFP_Ara | ||
| - | ** | + | ** pSB1K3_CFP_Lac |
| - | ** | + | ** pSB1K3_YFP_Lac |
* LB medium | * LB medium | ||
| Line 7,813: | Line 7,813: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Test Expression Backbones: pSB1K3_Ara/ | + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Test Expression Backbones: pSB1K3_Ara/Lac_CFP</h3> |
<b>Time:</b> 2011-09-10,11:00 <br> | <b>Time:</b> 2011-09-10,11:00 <br> | ||
| Line 7,821: | Line 7,821: | ||
<b>Aim:</b> | <b>Aim:</b> | ||
| - | prove of promotors ( | + | prove of promotors (Lac/Ara) |
<b>Materials/Methods</b> | <b>Materials/Methods</b> | ||
| Line 7,829: | Line 7,829: | ||
{| border="1" class="wikitable" | {| border="1" class="wikitable" | ||
| - | | '''time''' || '''time of grow [min]''' || '''OD600 Ara_CFP''' || '''OD600 Ara_CFP control''' || '''OD600 | + | | '''time''' || '''time of grow [min]''' || '''OD600 Ara_CFP''' || '''OD600 Ara_CFP control''' || '''OD600 Lac_CFP''' || '''OD600 Lac_CFP control''' || |
|- | |- | ||
| Line 7,893: | Line 7,893: | ||
** pSB1A3_YFP_Ara | ** pSB1A3_YFP_Ara | ||
| - | ** | + | ** pSB1A3_YFP_Lac |
** pSB1K3_CFP_Ara | ** pSB1K3_CFP_Ara | ||
| - | ** | + | ** pSB1K3_CFP_Lac |
| - | ** | + | ** pSB1K3_YFP_Lac |
** mdnA Lib-foc2 | ** mdnA Lib-foc2 | ||
| Line 8,189: | Line 8,189: | ||
** pSB1K3_CFP_Ara | ** pSB1K3_CFP_Ara | ||
| - | ** | + | ** pSB1K3_CFP_Lac |
* 10 ml LB medium | * 10 ml LB medium | ||
Revision as of 19:04, 20 September 2011
82th Labday 2011-09-01
Digest of pUP089 (vector)
Investigator: Niels, Katharina
Aim: generate vector for ligation and transformation to generate a libary
Materials/Methods:
- pub089 (A)
- 5 µl DNA (~4000ng)
- 0,5 µl Sfo I
- 0,5 µl Aat II
- 3 µl 10x Buffer 4 (Biolabs)
- 21 µl water
- total volume: 30µl
Purification
- by NucleoSpin Extract II Kit, protocol for PCR purification
| backseat driver asks: why do you use only 5 µl? what about gel purification? |
Further tasks:
- dephosphorylation
- ligation
- transformation
miniprep of overnight cultures - mdnDE, mdnABC
Investigator: Niels, Jessica
Aim: Isolate DNA from over night cultures from 31.08.2011 (Katharina)
Materials/Methods:
1.miniprep:
- over night culture
- mdnABC
- clone 3
- clone 4
- clone 5
- clone 6
- clone 7
- mdnDE
- clone 1
- clone 2
- clone 3
- clone 4
- clone 5
- using Kit NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- elution in 50µl Elution buffer
| backseat driver: almost there, but a real plasmid name would be better. |
2. Preparation of glycerol stocks:
- adding 300 µl glycerol to 700 µl culture
Result:
Further tasks:
- test-digest
Digest of miniprep - mdnDE, mdnABC
Investigator: Niels, Jessica
Aim: prove of isolated DNA from over night cultures from 2011-09-01 (Niels)
Materials/Methods:
- DNA (miniprep)
- SpeI
- EcoRI
- Buffer 4
- BSA
- Water
'1.Digest:
- DNA (µl) / Water (µl)
- mdnABC
- clone 3 -
- clone 4 -
- clone 5 -
- clone 6 -
- clone 7 -
- mdnDE
- clone 1 -
- clone 2 -
- clone 3 -
- clone 4 -
- clone 5 -
- 3µl Buffer 4 (Neblab)
- 0,3µl BSA
- 1 µl Spe I
- 1 µl EcoRI
- incubate 2h at 37°C
Further tasks:
- analyze by gel
Testdigest of miniprep - mdnDE, mdnABC
Investigator: Niels, Katharina
Aim: prove of plasmid
Materials:
- DNA (miniprep)
- SpeI
- EcoRI
- Buffer 4
- BSA
- Water
Digest:
:
- mdnABC
- 1µl DNA
- 2 µl Buffer 2 (Neblab)
- 0,5 µl HindIII
- 0,5 µl AvaI
- 16 µl Water
- incubate 2h at 37°C
- mdnDE
- 1µl DNA
- 2 µl 10x Buffer 4 (Neblab)
- 0,5µl HpaI
- 16,5 µl Water
- incubate 2h at 37°C
Further tasks:
Ligation of pUP089 and Lib-2
Investigators: Steffi, Niels, Katharina
Material
- purified pUP089 (vector)
- purified Lib-2 (insert)
| backseat driver: do these poor pieces of DNA have no date or other referral? |
Method
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- 6 µl vector
- 2 µl insert
- 1h @ RT
Further tasks:
- transformation
Transformation of library2
Investigators:Katharina, Steffi
Aim: Transformation of Ligation
Materials:
- competent E. coli cells (XL1-Blue, 2011-08-29)
- ligation products: Lib2 + ligation control (2011-09-01, Nie, Kat, Ste)
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 30 min on ice,
- heat shock 45 sec at 42°C,
- incubation 3 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Kan)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
- 2 plates w/ Kan: mdn-lib2 + control
Picking clones for overnight cultures of pSB1C3 + mdnABC and DE, respectively
Investigators:Steffi, Katharina
Aim: check colonies for correct plasmid
Materials:
- agar plates from 2011-08-31, (fridge):
- pSB1C3+mdnABC
- pSB1C3+mdnDE
- LB medium
- chloramphenicol (25 mg/ml)
Protocol:
- 6 x: 5 ml LB medium + 5 µl chloramphenicol
- pick colony from plate (from each plate 3 colonies)
- transfer to LB medium
- incubate over night in 37°C shaker (200 rpm)
Output:
- over night cultures:
- pSB1C3+mdnABC
- clone 1
- clone 2
- clone 3
- pSB1C3+mdnDE
- clone 1
- clone 2
- clone 3
Further tasks:
- miniprep
- test digest
Digestion of NgoMIV_TEV-Protease_iGEM_BamHI, NgoMIV PreScission-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV fragments and pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5 and pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Paul, Sascha, Stefan, Sebastian
| backseat driver: be more precise with your spelling of PreScission. |
Materials:
4 samples NgoMIV_TEV-Protease_iGEM_BamHI:
- TEV 3: 71,7 ng/µl
- TEV 7: 65,3 ng/µl
- TEV 10: 58,6 ng/µl
- TEV 16: 62,0 ng/µl
2 samples NgoMIV_PreScission-Protease_iGEM_BamHI
- P1: 91,5 ng/µl
- P2: 82,5 ng/µl
2 samples HindIII_iGEM_AraC_NgoMIV
- A1: 103,7 ng/µl
- A2: 127,4 ng/µl
pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (contains TEV cleavage site): 470 ng/µl
pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5 vector (contains PRE cleavage site): 270,2 ng/µl
| backseat driver: nice list, but dates would be really cool. |
Digestion protocol:
1: NgoMIV_TEV-Protease_iGEM_BamHI (4x, each sample 1x):
- 10µl NgoMIV_iGEM_TEV-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
2: NgoMIV_PreScission-Protease_iGEM_BamHI (2x, each sample 1x):
- 10µl NgoMIV_PreScission-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
3: HindIII_iGEM_AraC_NgoMIV (2x, each sample 1x):
- 10µl HindIII_iGEM_AraC_NgoMIV
- 1µl HindIII
- 1µl NgoMIV
- 5µl 10x buffer = NEB4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
4: pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5:
- 3µl pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5
- 1µl HindIII
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 39.5µl pure water
- =50µl
5: pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5:
- 5µl pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5
- 1µl HindIII
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 37.5µl pure water
- =50µl
-->The reaction was allowed to proceed for 2h at 37°C!
Ligation of NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI or NgoMIV_iGEM_TEV-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV into pJC354-NheI-143C-Xho_blaFL_GGH5 or pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Paul, Sebastian, Sascha, Stefan
Aim:
- 1. Triple-ligation of NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI (573bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-143C-Xho_blaFL_GGH5 vector (~4700bp) to create pUP_SG2_TorA_CS-PreScission_bla_AraC-PreSciss
ion
- 2.Triple-ligation of NgoMIV_iGEM_TEV_iGEM_BamHI (760bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (~4700bp)to create pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
Calculation of volumes to be used with: [http://www.gibthon.org/ligate.html ligation calculator] with 1:1 molar ratio
Materials:
PreScission: 2 reaction batches (we have two digested Pre-fractions)
1:
- 2,9 µL NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI fragment (573bp, 5,4ng/µl)
- 2 µL AraC fragment (12.6 ng/µl)
- 3,1 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector (30.8 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
2:
- 2,5 µL NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI (573bp, 6,6 ng/µl) fragment
- 2,2 µL AraC fragment
- 3,3 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector (30.8 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
- 1 controls: As 2 but with water instead of fragment
TEV: 4 reaction batches (we have three digested NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment fractions)
1:
- 1.8 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 5,8 ng/µl)
- 1.4 µL AraC fragment (12,6 ng/µl)
- 4.9 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (12,9 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
2:
- 2.4 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 3,7 ng/µl)
- 1.2 µL AraC fragment (12,6 ng/µl)
- 4.3 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (12,9 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
3:
- 3.2 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 2.5 ng/µl)
- 1.1 µL AraC fragment (12,6 ng/µl)
- 3.8 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (12,9 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
4:
- 2.4 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 3,9 ng/µl)
- 1.2 µL AraC fragment (12,6 ng/µl)
- 4.4 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (12,9 ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
| backseat driver: are you doing the same thing four times? why do you fractionate your DNA? |
- 1 control: As 4 but with water instead of fragment
Used method:
ligation at room temperatur for 1h
Further task: Transformation of XL1blue cells with ligation products
Edit:
Competent cells were transformed with the complete ligation batches and plated on Cm containing plates!
Results:
1: PreScission containing colnes, 10 colonies picked
2: TEV containing clones, 10 colonies picked
Note: "Rest" means, that after plating 100µl on a plate remaining cells in a tube were centrifugated, resuspended and plated again on a new plate.
plasmid pereperation and test digestion pPDV089 to control deletion of kanamycin gene, pARW089 containing only geneIII (no mdnA), pSB1C3 containing mdnA, geneIII or mdnA/geneIII
Investigators: Sandrina, Laura
Aim: control deletion of kanamycin gene in created pPDV089, control ligation of geneIII into PARW089, control ligation of mdnA, geneIII and mdnA/geneIII into pSB1C3
Method/Materials:
plasmid preperation:
protocol 5.1 of the NucleoSpin Plasmid Kit
test digestion:
- 5 clones from pPDV089_2S14 (ampicillin):
- 4 µl (132-256 ng/µl) vector DNA
- 0.5 µl NsiI
- 0.5 µl SpHI
- 2 µl buffer 2
- 13 µl water
incubate for 1 h at 37°C
- 5 clones from pARW089 with geneIII (kanamycin):
test digestion could not be done, because cells did not grow over night
- 5 clones from pSB1C3 with mdnA (chloramphenicol), 5 clones from pSB1C3 with geneIII (chloramphenicol), 5 clones from pSB1C3 with mdnA/geneIII (chloramphenicol):
- 4 µl vector DNA (58- 120 ng/µl)
- 2 µl buffer 3
- 0,2 µl BSA
- 0.5 µl XbaI
- 0.5 µl PstI
12.8 µl water
incubate for 1 h at 37°C
Results:
Loading of gels
| lane | Sample | Volume in µl | Expected insert size in bp |
| M | marker, DNA ladder mix Fermentas | ||
| 1 | geneIII in pSB1C3, clone 1 | 10 | ca. 500 |
| 2 | geneIII in pSB1C3, clone 2 | 10 | ca. 500 |
| 3 | geneIII in pSB1C3, clone 3 | 10 | ca. 500 |
| 4 | geneIII in pSB1C3, clone 4 | 10 | ca. 500 |
| 5 | geneIII in pSB1C3, clone 5 | 10 | ca. 500 |
| 6 | free | ||
| 7 | mdnA in pSB1C3, clone 1 | 12 | ca. 180 |
| 8 | mdnA in pSB1C3, clone 2 | 12 | ca. 180 |
| 9 | mdnA in pSB1C3, clone 3 | 12 | ca. 180 |
| 10 | mdnA in pSB1C3, clone 4 | 12 | ca. 180 |
| 11 | mdnA in pSB1C3, clone 5 | 12 | ca. 180 |
| 12 | pPDV089, clone 1 | 12 | ca. 1400 |
| 13 | pPDV089, clone 2 | 12 | ca. 1400 |
| 14 | pPDV089, clone 3 | 12 | ca. 1400 |
| 15 | pPDV089, clone 4 | 12 | ca. 1400 |
| 16 | pPDV089, clone 5 | 12 | ca. 1400 |
| 17 | free | ||
| 18 | mdnA-geneIII fusion gene in pSB1C3, clone 1 | 12 | ca. 700 |
| 19 | mdnA-geneIII fusion gene in pSB1C3, clone 2 | 12 | ca. 700 |
| 20 | mdnA-geneIII fusion gene in pSB1C3, clone 3 | 12 | ca. 700 |
| 21 | mdnA-geneIII fusion gene in pSB1C3, clone 4 | 12 | ca. 700 |
| 22 | mdnA-geneIII fusion gene in pSB1C3, clone 5 | 12 | ca. 700 |
Further tasks:
- sequncing of geneIII in pSB1C3, clone 3, mdnA-geneIII fusion gene in pSB1C3, clone 4 and mdnA in pSB1C3, clone 4
- phage display with pPDV089
- repeat ligation of geneIII in pARW089
overnight culture of picked E. coli clones transformed with pARW089 containing only geneIII (no mdnA)
Investigators: Sandrina, Laura
Aim: control ligation of geneIII into PARW089
Method/Materials:
- 4 clones from pARW089 with geneIII (kanamycin)
- 5 ml LB medium per clone
- storage over night at 37°C and 750 rpm
Further tasks:
- test digestion
Production of phages containing pPDV089
Investigators: Sandrina, Laura
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- first: amplification of cells containing pPDV089
- 50 ml DYT medium will be inoculated with the cells, so that OD600 = 0.1
- antibiotics tetracyclin and ampicillin should be geiven to the medium
- cells should be incubated 37 °C till OD600 = 0.3-0.5 is reached
- upcomming problem: cells died
Further tasks:
- repeat this procedure
83th Labday 2011-09-02
Miniprep of overnight cultures from ligation of pSB1C3 + mdnABC and mdnDE, respectively
Investigators: Vanessa, Katharina
Time: 2011-09-02
Aim: DNA for sequencing and confirmation of insert
Materials:
- 6 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
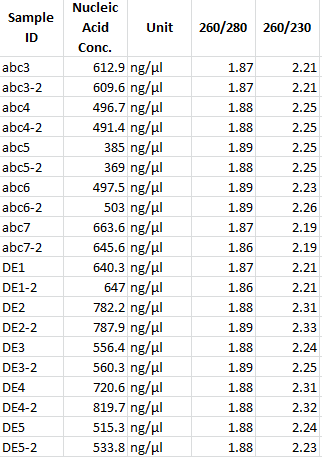
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnABC clone 1 | 745.7 |
| mdnABC clone 2 | 1121.8 |
| mdnABC clone 3 | 974.6 |
| mdnDE clone 1 | 873.2 |
| mdnDE clone 2 | 665.5 |
| mdnDE clone 3 | 665.7 |
test digest of pSB1C3 + mdnABC and DE, respectively
Time: 2011-09-02
Investigators: Steffi, Katharina
Aim: prove of Insert (mdnABC or mdnDE)
Materials:
Digestion protocol for pSB1C3+mdnABC
- 1 µl DNA
- 0.3 µl BSA
- 1 µl HincII
- 3 µl 10x buffer = NEB 3
- 24.7 µl pure water
- 37°C for 1h
Digestion protocol pSB1C3+mdnDE
- 1 µl DNA
- 1 µl HpaI
- 3 µl 10x buffer = NEB 4
- 25 µl pure water
- 37°C for 1h
Production of one 1 %
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker | ||
| 1 | mndABC clone 1 | 12 | 486, 1482, 2884 |
| 2 | mdnABC clone 2 | 12 | 486, 1482, 2884 |
| 3 | mdnABC clone 3 | 12 | 486, 1482, 2884 |
| 4 | mdnDE clone 1 | 12 | 1329, 3825 |
| 5 | mdnDE clone 2 | 12 | 1329, 3825 |
| 6 | mdnDE clone 3 | 12 | 1329, 3825 |
Results:
Conclusions:
- test digest seems to be correct for:
- lane 2, 5 and 6
Further task:
- send positive plasmids for sequencing
Colony PCR of plated cells from 01.09.2011 (see entry from 01.09.2011 for picked colonies)
Investigators: Sascha, Paul, Sebastian
Aim:
Proove whether triple ligation of TEV and PreScission into backbone was succesful.
Used Primer:
Tev:
f_AraC_HindIII_iGEM, r_TEV_iGEM_BamHI
PreScission:
f_AraC_HindIII_iGEM, r_Pre_iGem:BamHI
Method:
- 2,5µl of each primer (10µM) = 5µl
- 5µl 10y polymerase buffer
- 2µl 25mM MgCl2
- 1µl dNTP mix
- 0,5µl Taq-Polymerase (GenAxxon)
- 36,5 µl H2O
=50µl
Colonies were picked with a 20µl tip, dipped into PCR batch, and then plunged into 1ml LB to grew cells from picked colony (from these 1ml batches overnight cultures will be inoculated in case of positive clones).
- 10 colonies were picked for TEV and 10 colonies for PreScission protease (see entry from 01.09.2011) = 20 PCR batches.
PCR Program:
Initial denat = 3min 94°C
25x
denat: 2min 10sec 94°C
anneal: 2min 10sec 70°C
extend: 2min 10sec 72°C
final extend: 10min 72°C
- The PCR products were resolved on a 1% analytical agarose gel
Expected fragments:
Tev+AraC ~ 2033 bp
Pre+AraC ~ 1846 bp
Results:
- Based on the results from the agarose gel overnight precultures from the 1 ml batches were prepared.
Following clones were used:
Tev:
T2, T4, T5, T6, T7, T8, T9, T10
PreScission:
P1, P3, P4, P6; P8, P9, P10
over night cultures of pSB1C3+mdnABC clone 2 and P1 for screening team
time: 2011-9-2, 18:00
Investigators: Nadine, Jessica
Aim: preparation of gycerol stock from pSB1C3+mdnABC transformed cells (cultures were placed instable in the incubator)
Materials:
- over night culture from Katharina (2011-9-1)
- pSB1C3+mdnABC clone 2
- over night culture from Paul (2011-9-2):
- P1
- LB medium
- chloramphenicol (25 mg/ml)
Protocol:
- 2 x: 10 ml LB medium + 10 µl chloramphenicol
- use left cell suspension for inoculation
- transfer to LB medium
- incubate over night in 37°C shaker (200 rpm)
Output:
- over night cultures:
- pSB1C3+mdnABC
- clone 3
- P1 for screening team
Further tasks:
- mdnABC: glycerol stocks
plate transformation of pUP089A and B (XL1-blue)
time: 2011-9-2, 17:00
Investigators: Nadine, Jessica
Aim: get new pUP089 vector that is not contaminated, check new plasmid for antibiotic resistance
Materials:
- Trafo from Jessica (2011-9-2)
- pUP089 A
- pUP089 B
- Agar plates w/:
- 100 µg/ml Cm (2x)
- 100 µg/ml Amp (1x)
- 100 µg/ml Kan (2x)
Protocol:
- centrifuge incubated cells
- resuspend in 16 µl LB
- plate 50 µl
- incubate over night in 37°C
Output:
- Agar plates:
- Cm pUP089 A
- Cm pUP089 B
- Kan pUP089 A
- Kan pUP089 B
- Amp pUP089 A
Further tasks:
- check plates
- over night cultures
PCR of mdnA with N-terminal myc tag (mcyN)
Investigators: Jessica
Time: 2011-09-02, 14:00
Material:
- pARW089 (~8 ng/µl)
- dNTPs
- primer 23, 24
- HF Phusion Buffer 5x
- Phusion Polymerase
Method:
- 1 µl pARW089
- 1 µl dNTPs (10 mM each)
- 2.5 µl forward primer (10µM)
- 2.5 µl reverse primer (10µM)
- 10 µl HF Phusion Buffer 5x
- 0.5 µl Phusion Polymerase
- 32.5 µl water
- total 50 µl
- edited programm IGBIO2
| Step | Temperature | Time | |
|---|---|---|---|
| Hot Start | 98°C | Hold | |
| Initial denaturation | 98°C | 30 sec | |
| Denaturation | 98°C | 10 s | 10x |
| Annealing | 58°C | 20 s | |
| Extension | 72°C | 20 s | |
| Denaturation | 98°C | 10 s | 20x |
| Annealing | 72°C | 20 s | |
| Extension | 72°C | 20 s | |
| Final extension | 72°C | 10 min | |
| 4°C | Hold |
Result:
- PCR product mycN (stored in fridge, violett reck)
NOTE: PCR mycC done in july is stored at -20°C in blue PCR box (gel purified)
Further Tasks:
- gel electrophoresis
- PCR clean up
- restriction enzyme digestion
- ligation
- transformation
Transformation of XL1blue-cells with pPDV089
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- addition of 0,3 µl ligated vector pPDV089 from clone 2S14 (after plasmid preperation) to competent XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/ml ampicillin
- storage over night at 37°C
Further tasks:
- over night culture, amplification of cells for phage display, test digestion
Send pSB1C3 vectors, containing geneIII, mdnA and geneIII/mdnA fusion gene, for sequencing
Investigators: Sandrina
Aim: control if mdnA, geneIII and mdnA/geneIII fusion gene were fully ligated in pSB1C3 to generate biobricks
Method/Materials:
- sending clones geneIII in pSB1C3, clone 3, mdnA-geneIII fusion gene in pSB1C3, clone 4 and mdnA in pSB1C3, clone 4 to GATC
- 20 µl, 50-100 ng/µl
- primer:
Further tasks:
- sequence alignment
Plasmid preperation and test digestion of pARW089 containing only geneIII (no mdnA)
Investigators: Sandrina
Aim: control if ligation of geneIII in pARW089 worked
Method/Materials:
plasmid preperation:
- protocol 5.1 of the NucleoSpin Plasmid Kit
- test digestion:
- 4 clones from pARW089 with geneIII:
- 4 µl (132-256 ng/µl) vector DNA
- 0.5 µl SfoI
- 0.5 µl AatII
- 2 µl buffer 4
- 13 µl water
- incubate for 1 h at 37°C
- glycerol stocks of clone 1, 2 and 4 (stored at -80°C)
Results:
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker, DNA ladder mix Fermentas | ||
| 1 | geneIII in pARW089, clone 1 | 10 | ca. 10000 and 550 |
| 2 | geneIII in pARW089, clone 2 | 10 | ca. 10000 and 550 |
| 3 | geneIII in pARW089, clone 3 | 10 | ca. 10000 and 550 |
| 4 | geneIII in pARW089, clone 4 | 10 | ca. 10000 and 550 |
Further tasks:
- sequence vector
84th Labday 2011-09-03
PCR-clean up of mdnA with N-terminal myc tag (mcyN) (Jessica 2011-09-02)
Investigators: Nadine, Katharina
Time: 2011-09-03, 9:00
Material:
- PCR product of mycN
- NucleoSpin Extract II Kit from Macherey-Nagel
- elution in 50 µl
Result:
- concentration of product:
- 40.0 ng/µl
Restriction enzyme digestion of mycN, mycC and pSB1C3
Investigators: Nadine, Katharina
Time: 2011-09-03, 10:00
Material:
- purified PCR products
- mycC (4.8 ng/µl (2011-07-05 Jessica))
- mycN (40.0 ng/µl)
- NEB Buffer 4
- XbaI
- PstI
- BSA
- digestion of mycN and mycC
- 30 µl DNA
- 5 µl Buffer 4
- 1.5 µl XbaI
- 2.0 µl PstI
- 0.5 µl BSA
- 12 µl water
- digestion of pSB1C3
- 6 µl DNA (235.0 ng/µl (2011-06-22))
- 3 µl Buffer 4
- 1.5 µl XbaI
- 2.0 µl PstI
- 0.5 µl BSA
- 11.5 µl water
- concentration after digestion
- mycC: 4.2 ng/µl
- mycN: 15.2 ng/µl
- pSB1C3: 16.4 ng/µl
Ligation of mycC and mycN, respectively, and pSB1C3
Time: 2011-09-03, 17:30
Investigators: Nadine, Katharina
Materials
- T4 DNA Ligase Buffer (Fermentas)
- T4 DNA Ligase
- purified samples of Lib2 and pUP089
Method mycC
- 1µl T4 DNA Ligase Buffer
- 1µl T4 DNA Ligase
- 5µl pSB1C3
- 3µl purified mycC
- also preparing ligation control (3µl water instead of mycC)
- incubation for 1h at roomtemperature
Method mycN
- 1µl T4 DNA Ligase Buffer
- 1µl T4 DNA Ligase
- 7µl pSB1C3
- 1µl purified mycN
- incubation for 1h at roomtemperature
Transformation of XL1 with mycC/mycN in pSB1C3
Time: 2011-09-03, 19:00
Investigators: Nadine, Katharina
Materials:
- XL1-Blue
- ligation products: pSB1C3+mycC and pSB1C3+mycN
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 90 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Kan)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
- 2 plates with Cm: mycC, mycN
- 1 plate with Cm: control myc
Oligo-Fillin for mdnA-Library 2 (repetition)
Time: 2011-09-03, 11:00
Investigators: Nadine, Katharina
Materials
- Primer, 25 µM :
- # 74: o_foc_library_2
- Klenow-Buffer 10X
- Klenow Fragment
- dNTPs
- water
Protocol:
- Reaction mix 1
- 1 µl fw-Oligonucleotide (#76)
- 1 µl rev-Oligonucleotide (#74)
- 2 µl dNTPs (10 mM)
- 2 µl Klenow-Buffer
- 0.5 µl Klenow Fragment
- 14 µl water
- total volume: 20.5 µl
- Reaction mix 2
- 2 µl fw-Oligonucleotide (#76)
- 2 µl rev-Oligonucleotide (#74)
- 2 µl dNTPs (10 mM)
- 2 µl Klenow-Buffer
- 0.5 µl Klenow Fragment
- 12 µl water
- total volume: 20.5 µl
- reaction mix 3
- 3 µl fw-Oligonucleotide (#76)
- 3 µl rev-Oligonucleotide (#74)
- 2 µl dNTPs (10 mM)
- 2 µl Klenow-Buffer
- 0.5 µl Klenow Fragment
- 10 µl water
- total volume: 20.5 µl
2. PCR program
- name: Fillin
- 3 min 94 °C
- 0.3°C per s (94°C-37°C)
- addition of 0.5 µl Klenow Fragment
- press enter
- 1hr 37°C
Agarose gel:
1 %: 0.5 g in 50 ml TAE
Results:
- Output:
- mdnA-Lib2 1µl
- mdnA-Lib2 2µl
- mdnA-Lib2 3µl
Further tasks:
- Gel purification
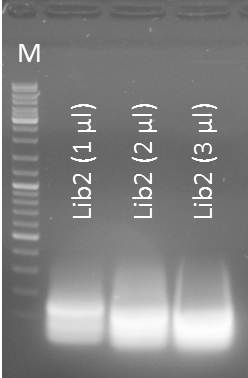
Gel purification of Oligo-Fillin for mdnA-Library 2 (repetition)
Time: 2011-09-03, 13:00
Investigators: Nadine, Katharina
Materials
- Macherey Nagel - NucleoSpin Extract II, protocol for DNA extraction from agarose gels
- Eluation with 50µl NE-Buffer
NanoDrop: Concentrations
- Lib2 (1 µl) purified: 13.8 ng/µl
- Lib2 (2 µl) purified: 14.9 ng/µl
- Lib2 (3 µl) purified: 9.0 ng/µl
Restriction enzyme digestion of Oligo-Fillin for mdnA-Library 2 (repetition) and pUP089
Time: 2011-09-03, 13:00
Investigators: Nadine, Katharina
Materials
- purified Lib2 samples
- pUP089 (2011-06-22)
- AatII
- NEB Buffer 4
Method
Digestion protocol for Lib2
- 50 µl purified Lib2 sample
- 2.0 µl AatII
- 6 µl Buffer 4
- 2.0 µl water
- 37°C for 1 h
Digestion protocol for pUP089:
- pUP089
- NEB Buffer 4
- SfoI
- AatII
- digestion protocol
- 4 µl DNA
- 3µl Buffer 4
- 1.5 µl AatII
- 1.5 SfoI
- 20 µl water
- incubation @ 37°C for 4 h
- control digestion 1
- 2 µl DNA
- 3 µl Buffer 4
- 1.5 µl AatII
- 23.5 µl water
- incubation @ 37°C for 4 h
- control digestion 2
- 2 µl DNA
- 3 µl Buffer 4
- 1.5 µl SfoI
- 23.5 µl water
- incubation @ 37°C for 1h
Loading of gels
| lane | Sample | ||
| M | marker, DNA ladder mix Fermentas | ||
| 1 | Lib2 (1µl) digested | ||
| 2 | Lib2 (2µl) digested | ||
| 3 | Lib2 (3µl) digested | ||
| 4 | pUP SfoI/AatII | ||
| 5 | pUP AatII (control) | ||
| 6 | pUP SfoI (control) |
- fragments were excised and purified using Macherey-Nagel Nucleo SpinII Extract Kit
- concentrations after purification
- Lib2 (1µl): 6.8 ng/µl
- Lib2 (2µl): 4.7 ng/µl
- Lib2 (3µl): 4.9 ng/µl
- pUP089: 14.9 ng/µl
Ligation of Lib2 (repetition) and pUP089
Time: 2011-09-03, 17:30
Investigators: Nadine, Katharina
Materials
- T4 DNA Ligase Buffer (Fermentas)
- T4 DNA Ligase
- purified samples of Lib2 and pUP089
Method
- 1µl T4 DNA Ligase Buffer
- 1µl T4 DNA Ligase
- 7µl pUP089
- 1µl Lib2 (1µl)
- also preparing ligation control (1µl water instead of Lib2)
- incubation for 1h at roomtemperature
Transformation of XL1 with Lib2 in pUP089
Time: 2011-09-03, 19:00
Investigators: Nadine, Katharina
Materials:
- XL1-Blue
- ligation products: pUP089+Lib2
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 90 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Kan)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
- 10 plates with Kan: Lib2
- 1 plate with Kan: control Lib2
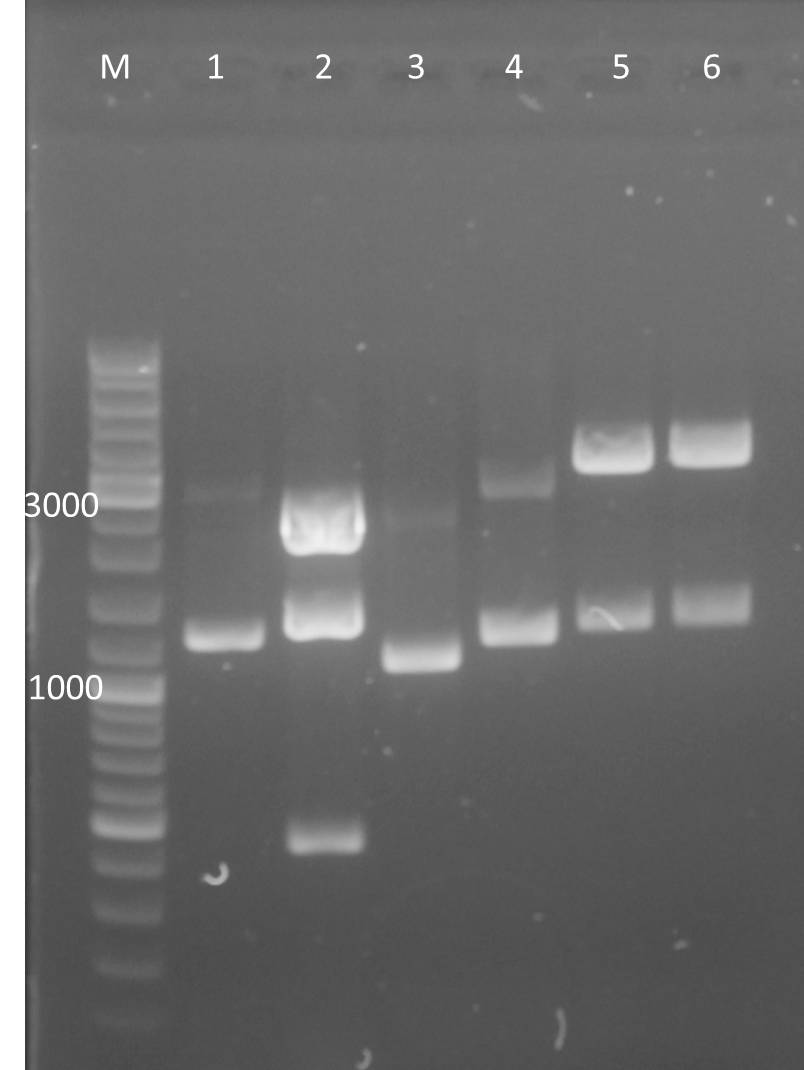
Transformation of XL1 with K3 and A3 expression backbones
Time: 2011-09-03, 19:00
Investigators: Nadine, Katharina
Materials:
- XL1-Blue
- DNA of:
- pSB1A3_YFP_Ara clone A
- pSB1A3_YFP_Lac clone B
- pSB1K3_CFP_Ara clone A
- pSB1K3_CFP_Lac clone B
- pSB1K3_YFP_Lac clone C
Method:
- addition of 2 µl DNA to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Kan/Amp)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Picking clones for overnight cultures of pSB1C3 + DE clone 2 and clone 3
Investigators:Sebastian, Katharina
Time: 2011-09-04, 16:00
Materials:
- liquid cultures of mdnDE clone 2 and mdnDE clone 3
- LB medium
- chloramphenicol (25 mg/ml)
Method:
- inoculate 1 ml of liquid culture to 4 ml LB medium+ Cm
- incubate over night in 37°C shaker (200 rpm)
Further tasks:
- miniprep
- sending for sequencing
overnight culture of picked E. coli clones transformed with of XL1blue-cells with pPDV089
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- 3 clones from pPDV089, clone 2S14, (ampicillin and tetracyclin)
- 5 ml LB medium per clone
- storage over night at 37°C and 750 rpm
Further tasks:
- amplification for phage display and test digestion
85th Labday 2011-09-04
Production of phages containing pPDV089 in XL1 blue cells
Investigators: Sandrina, Katharina, Nadine
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- first step: amplification of cells containing pPDV089( clone: 2S14):
- 50 ml DYT medium will be inoculated with the cells, so that OD600 = 0.1
- add antibiotics tetracyclin and ampicillin to the medium
- cells should be incubated 37 °C till OD600 = 0.3-0.5 (here: 0.332) is reached
- second step: infection with helper phages
- add helper phages 10^11 phages/50 ml (...)
- incubate for 10 min at 37°C (without shaking!)
- add 0,5 mM IPTG
- incubate 50 min at 28°C and rpm
- add 70 µg/ml kanamycin and incubate for 5 h at 28°C (...)
- third step: phage purification
- centrifuge cell culture at 5000 x g/ 15 min
- fill supernatant in a new 50 ml falcon and centrifuge again (5000 g/15 min)
- 40 ml of the supernatant with 8 ml PEG-NaCl (20% (w/v) PEG-8000, 2,5 M NaCl)
- incubate over night at 4°C
Further tasks:
- go on with phage purification
Miniprep of cells picked from plates on 02.09.2011 and inoculated into ON precultures
Investigators: Sandrina, Katharina
Time: 2011-09-04, 11:00
1. Miniprep:
- 15 overnight cultures (pUP SG3- pUP SG17)
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| pUP SG3 | 1410.2 |
| pUP SG4 | 1146.2 |
| pUP SG5 | 1289.7 |
| pUP SG6 | 940.1 |
| pUP SG7 | 1418.5 |
| pUP SG8 | 1244.4 |
| pUP SG9 | 1287.6 |
| pUP SG10 | 1188.7 |
| pUP SG11 | 1276.1 |
| pUP SG12 | 1193.9 |
| pUP SG13 | 1255.1 |
| pUP SG14 | 1474.7 |
| pUP SG15 | 1143.5 |
| pUP SG16 | 1268.0 |
| pUP SG17 | 1218.7 |
Miniprep of overnight cultures from ligation of pSB1C3+mdnDE clone 2 and clone3
Investigators: Sandrina, Katharina
Time: 2011-09-04, 11:00
Material:
- 2 overnight cultures (mdnDE clone 2 and mdnDE clone 3)
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
Results:
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnDE clone 2 | 601.9 |
| mdnDE clone 3 | 571.7 |
Further Tasks:
- sending for sequencing
Transformation of XL1 with mdnBC in pSB1C3
Time: 2011-09-04, 14:00
Investigators: Nadine, Katharina
Materials:
- XL1-Blue
- ligation products: pSB1C3+mdnBC (Niels 2011-08-24)
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Cm)
- storage over night at 37°C
- no ligation control available
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Control experiment: Transformation of XL1 with pARW089 and pARW071, respectively
Time: 2011-09-04, 14:00
Investigators: Nadine, Katharina
Materials:
- XL1-Blue
- DNA of pARW089 and pARW071 (2011-05-24)
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Cm)
- storage over night at 37°C
- no ligation control available
Picking clones for overnight cultures of pSB1C3+mycC and pSB1C3+mycN
Investigators:Katharina
Time: 2011-09-04, 16:00
Materials:
- agar plates of pSB1C3+mycC and pSB1C3+mycN
- LB medium
- chloramphenicol (25 mg/ml)
Method:
- picking 5 clones for each construct
- incubate over night in 37°C shaker (200 rpm)
Output:
- 5 liquid cultures of mycC (1-5)
- 5 liquid cultures of mycN (1-5)
Further tasks:
- miniprep
- sending for sequencing
Picking clones for overnight cultures of pARW089 and pARW071
Investigators:Katharina
Time: 2011-09-04, 16:00
Materials:
- Glycerolstocks of pARW089 and pARW071
- LB medium
- Kan
Method:
- preparing one liquid culture for each construct
- incubate over night in 37°C shaker (200 rpm)
Further tasks:
- preparation of 400ml culture for HPLC analysis on 2011-09-05
86th Labday 2011-09-05
Test digest of isolated plasmids (4.9.2011) from clones that were positive in colony PCR (2.09.2011)
Investigators: Paul, Stefan, Sebastian
Aim: Prove whether plasmids contain all expected inserts.
Method: Digestion of plasmids with HincII. Produces 3 fragments from both, the PreScission and the TEV containing plasmids.
- Expected fragments:
- Tev: 550 bp, 2700 bp, 3400 bp
- Pre: 550 bp, 2300 bp, 3600 bp
- 5µl 10x buffer 4 (NEB)
- 0,5µl 100x BSA
- 1µl Sample
- 0,5µl HincII Enzyme (NEB)
- 43µl water
- 15 reaction batches, 7 for PreScission, 8 for TEV:
following colnes were used:
2 pUP_SG3_ssTorA_CS-Pre_blaFL_AraC-Pre1
3 pUP_SG4_ssTorA_CS-Pre_blaFL_AraC-Pre3
4 pUP_SG5_ssTorA_CS-Pre_blaFL_AraC-Pre4
5 pUP_SG6_ssTorA_CS-Pre_blaFL_AraC-Pre6
6 pUP_SG7_ssTorA_CS-Pre_blaFL_AraC-Pre8
7 pUP_SG8_ssTorA_CS-Pre_blaFL_AraC-Pre9
8 pUP_SG9_ssTorA_CS-Pre_blaFL_AraC-Pre10
11 pUP_SG10_ssTorA_CS-TEV_blaFL_AraC-TEV2
12 pUP_SG11_ssTorA_CS-TEV_blaFL_AraC-TEV4
13 pUP_SG12_ssTorA_CS-TEV_blaFL_AraC-TEV5
14 pUP_SG13_ssTorA_CS-TEV_blaFL_AraC-TEV6
15 pUP_SG14_ssTorA_CS-TEV_blaFL_AraC-TEV7
16 pUP_SG15_ssTorA_CS-TEV_blaFL_AraC-TEV8
17 pUP_SG16_ssTorA_CS-TEV_blaFL_AraC-TEV9
18 pUP_SG17_ssTorA_CS-TEV_blaFL_AraC-TEV10
Results:
competent cells - ER2738
Investigator: Katharina, Niels, Sandrina, Steffi
Aim: produce competent cells
Materials/Methods:
| TFB I | 1000ml | 200ml |
| 100mM Rubidium Chloride | 12.1 | 2.42g |
| 30mM Potassium Acetate | 2.944 | 0.59g |
| 10mM Calcium Chloride | 1.47 | 0.29g |
| 15% w/v Glycerol (87%) | 150 | 34.5g |
Adjust pH to 5.8 with acetic acid
Filter sterilize the solution
| TFB II | 500ml | 100ml |
| 50mM Rubidium Chloride | 0.6 | 0.121g |
| 10mM MOPS | 1.05 | 0.210g |
| 75mM Calcium Chloride | 5.51 | 1.100g |
| 15% w/v Glycerol (87%) | 75 | 17.24g |
Adjust pH to 7.0 with KOH
Filter sterilize the solution
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- Prepare 80 Eppis (1,5?l)
- get liquid nitrogen
- prepare 5 ml LB-Medium with the specific antibiotic (for ER2738: Tet), inoculate and incubate over night
- prepare 200 ml LB-Medium with the specific antibiotic, inoculate with 2 ml of the over-night-culture
- grow while shaking at 37°C, 190 rpm to an OD600 at 0,4-0,6
- keep cell suspension in sterile falcons (50 ml) 10 min on ice, then centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend on ice with 40 ml icecold TFB I and keep 10 min on ice
- centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend pellet in 8 ml TFB II
- aliquot in Eppis: 50?l per tube and store immediately at liquid nitrogen and afterwards at -80 °C
Results:
- 80 tubes ER2738 for transformation(50 µl competent cells at -80°C)
Further tasks:
check by transformation and check the resistance on agar plates with different antibiotics
check resistance of competent cells - E.coli ER2738
Investigator: Niels, Katharina, Sandrina, Steffi
Aim: check resistance of competent ER2738-cells on agar plates with different antibiotics
Materials/Methods:
- competent ER2738-cells from 2011-09-05 (Niels, Katharina, Sandrina, Steffi)
- LB-plates with Tetracycline, Chloramphenicol, Kanamycine, Ampicillin, without antibiotics
- plating 50 µl on agar plates
- incubate at 37°C over night
Further tasks:
control agar plates
Miniprep of overnight cultures of pSB1C3+mycC and pSB1C3+mycN
Investigators:Nadine, Nicole, Katharina
Time: 2011-09-04, 9:00
Materials:
- liquid culture of pSB1C3+mycC and pSB1C3+mycN
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mycN clone 1 | 400.6 |
| mycN clone 2 | 343.0 |
| mycN clone 3 | 395.4 |
| mycN clone 4 | 618.0 |
| mycN clone 5 | 551.1 |
| mycC clone 1 | 519.7 |
| mycC clone 2 | 453.3 |
| mycC clone 3 | 631.5 |
| mycC clone 4 | 689.5 |
| mycC clone 5 | 897.3 |
Further tasks:
- test digestion for confirmation
- sending for sequencing
Cultures of pARW089 and pARW071 for HPLC-Analysis
Investigators:Katharina, Nadine, Nicole, Steffi
Materials:
- overnight cultures of pARW089 and pARW071 from 2011-09-04 (Katharina)
- LB medium
- Kan
Method:
- preparing one liquid culture for each construct with Kanamycine
- incubate for 6 hrs in 37°C shaker (190 rpm)
Further tasks:
- HPLC analysis
Oligo-Fillin for mdnA-Library 2 (repetition)
Time: 2011-09-05, 10:00
Investigators: Nadine, Katharina, Jessica, Nicole, Steffi
Aim: Repetition of fillin reaction for library with new fillin program
Materials
- Primer, 25 µM :
- # 76
- # 74
- Klenow-Buffer 10X
- Klenow Fragment
- dNTPs
- water
Protocol:
- Reaction mix (50 µl)
- 3 µl fw-Oligonucleotide (#76)
- 3 µl rev-Oligonucleotide (#74)
- 5 µl dNTPs (10 mM)
- 5 µl Klenow-Buffer
- 34 µl water
- total volume: 50 µl
- total volume: 50 µl
(2. Fillin program
- name: Fillin
- 3 min 94 °C
- 0.3°C per s (94°C-37°C)
- addition of 0.5 µl Klenow Fragment
- press enter
- 1hr 37°C)
Results:
- Output:
- mdnA-Lib2, Nad, 2011-9-5
Further tasks:
- digest w/ AatII
- ligation w/ pUP089 (digested w/ AatII and SfoI)
- transformation
Control restriction enzyme digestion of pSB1C3_mdnA_mycC and pSB1C3_mdnA_mycN
Investigators: Nadine, Nicole, Katharina, Steffi
Aim: Confirmation of the insertion of mdnA with myc-tag (N-terminal and C-terminal) in pSB1C3 vector
Time: 2011-09-05, 10:00-11:30
Material:
- clones:
- pSB1C3_mdnA_mycC, clone 1, Katharina, 2011-09-05
- pSB1C3_mdnA_mycC, clone 2, Katharina, 2011-09-05
- pSB1C3_mdnA_mycC, clone 3, Katharina, 2011-09-05
- pSB1C3_mdnA_mycC, clone 4, Katharina, 2011-09-05
- pSB1C3_mdnA_mycC, clone 5, Katharina, 2011-09-05
- pSB1C3_mdnA_mycN, clone 1, Katharina, 2011-09-05
- pSB1C3_mdnA_mycN, clone 2, Katharina, 2011-09-05
- pSB1C3_mdnA_mycN, clone 3, Katharina, 2011-09-05
- pSB1C3_mdnA_mycN, clone 4, Katharina, 2011-09-05
- pSB1C3_mdnA_mycN, clone 5, Katharina, 2011-09-05
- DraI (Fermentas)
- Tango buffer (Fermentas)
Method:
Reaction mix
- 2 µl DNA
- 2 µl Tango buffer
- 2 µl DraI (cuts three times)
- 14 µl H2O
Reaction conditions
- 37°C, 2 hours
Expected results:
- four fragments for each plasmid
- fragment 1: 771 bases
- fragment 2: 692 bases
- fragment 3: 19 bases
- fragment 4: 1114 bases
- pSB1C3_mdnA_mycC (four fragments)
- fragment 1: 804 bases
- fragment 2: 692 bases
- fragment 3: 19 bases
- fragment 4: 1082 bases
Conclusion:
Output: clone_name, cell type, stored fridge/freezer/-80°C; model saved as name in folder
PCR: mdnB, mdnABCDE
Time: 2011-09-05
Investigators: Nicole, Nadja
Materials
- vector: pARW089 (8,6 ng/µl), pARW071 (6,3 ng/µl), pARW089_T7 (8,6 ng/µl),
- 1-Phusion HF Polymerase, NEB
- 1-Phusion HF Buffer
- 2-PFU- Ultra HF Polymerase, NEB
- 2-PFU- Ultra HF Buffer, NEB
- dNTPs
- water
- Primer:
| # | Primer |
|---|---|
| 58 | pf_mdnB_EcoRI_NotI_XbaI 12.08. |
| 81 | pr_mdnB_SpeI_NotI_PstI_30.08. |
| 84 | pf_mdnABCDE89_EcoRI_NotI_XbaI |
| 82 | r_mdnABCDE_iGEM |
Protocol:
- mdnB
- 1 µl pARW089 (diluted 1:100)
- 1 µl dNTPs (10 mM)
- 2.5 µl Primer forward (10 µM)- 81
- 2.5 µl Primer backward (10 µM)- 58
- 10 µl Phusion buffer HF
- 0.5 µl Phusion HF Ploymerase (2U/µl)
- 32.5 µl water
- total volume: 50 µl
- mdnABCDE
- 1 µl pARW089, 1 µl pARW089_T7, 1,3 µl pARW071
- 1 µl dNTPs (10 mM)
- 1 µl 2-PFU- Ultra HF Polymerase, NEB
- 5 µl 2-PFU- Ultra HF Buffer, NEB
- 40 µl (pARW089 pARW089_T7) water, 39,7 µl (pARW071) water
- 1 µl Primer forward (10 mM) - 84
- 1 µl Primer backward (10 mM)- 82
2. PCR programs
- IGMDNBB for mdnB
- first steps: 10x
- second steps: 20x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 98°C | 10 s |
| Annealing | 64°C | 20 s |
| Elongation | 72°C | 25 s |
| DenaturationII | 98°C | 10 s |
| AnnealingII | 72°C | 20 s |
| ElongationII | 72°C | 30 s |
| Final Elongation | 72°C | 10 min |
- IGMDNAB
- first steps: 10x
- second steps: 20x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 98°C | Hold |
| Initial denaturation | 98°C | 30 sec |
| Denaturation | 98°C | 10 s |
| Annealing | 69°C | 20 s |
| Elongation | 72°C | 2 min 10 s |
| DenaturationII | 98°C | 30 s |
| AnnealingII | 72°C | 10 s |
| ElongationII | 72°C | 2min 30 s |
| Final extension | 72°C | 10 min |
Results:
- M: DNA ladder mix (Fermentas)
- 1: mdnABCDE from pARW089, exp. size: ~ 6500 bp
- 2: mdnABCDE+T7 from pARW089, exp. size: ~ 6500 bp
- 3: mdnABCDE from pARW071, exp. size: ~ 6500 bp
- 4: -
- 5: mdnB, exp. size: ~1000 bp
- Output:
- mdnB worked out
- mdnABCDE did not work out
Different overnight cultures
Investigators: Jessica, Nadja
Time: 2011-09-04, 18:00
Materials:
- glycerol stocks of pARW089 and pARW071
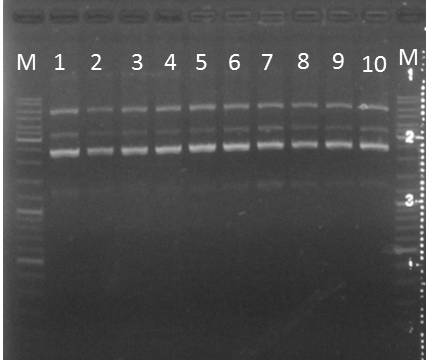
- plates of pSB1C3 + mdnBC (from 2011-09-04), pUP089 A and B, pSB1A3_Ara_YFP, pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP, pSB1K3_Lac_YFP (from 2011-09-03)
- LB medium
- Kan, Amp, Cm
Method:
- number of tubes: 5x mdnBC, 4x pUP089, 1x for the others
- incubate over night in 37°C shaker (200 rpm)
Further tasks:
- pSB1C3 + mdnBC: test digest
- pUP089: miniprep and glycerol stocks?
- expression backbones: glycerol stocks, preculture for expression test
- pARW089 and pARW071: cultures for HPLC
Purification of phages containing pPDV089
Investigators: Sandrina, Sabine
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- after over night incubation:
- centrifuge precipitaed phages: 5000 x g/ 45 min
- discard supernatant
- centrifuge again, 5000 x g/ 5 min
- remove supernatant carefully
- take pellet in 1 ml TBS
- move in 1.5 ml Eppi
- centrifuge again: 17000 x g/10 min
- supernatant in a new Eppi with 200 µl PEG-NaCl (mix!)
- incubate on ice for 60 min
- centrifuge precipitated phages: 17.000 x g/ 10 min (4°C)
- resuspend pellet in 300 µl TBS
- centrifuge:17 000 x g/ 10 min (4°C)
- supernatant in a fresh eppi= purified phages
Further tasks:
- measure concentration of phages
Glycerol stocks of phages containing pPDV089
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- 300 µl phages in TBS
- 750 µl 86 % glycerol
- store at -80°C
Further tasks:
- measure concentration of phages
Concentration of phages containing pPDV089 measured by nanodrop
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Results:
- OD 269: 0,095
- OD 320: 0,003
- phages per ml:
Ph/ml = (Abs 269- Abs 320) * 6*10^16/ bp (phagemid)
- bp (phagemid) = 10945
Ph/ml= 5*10^11
Further tasks:
- measure concentration of phages by serial dilution
87th Labday 2011-09-06
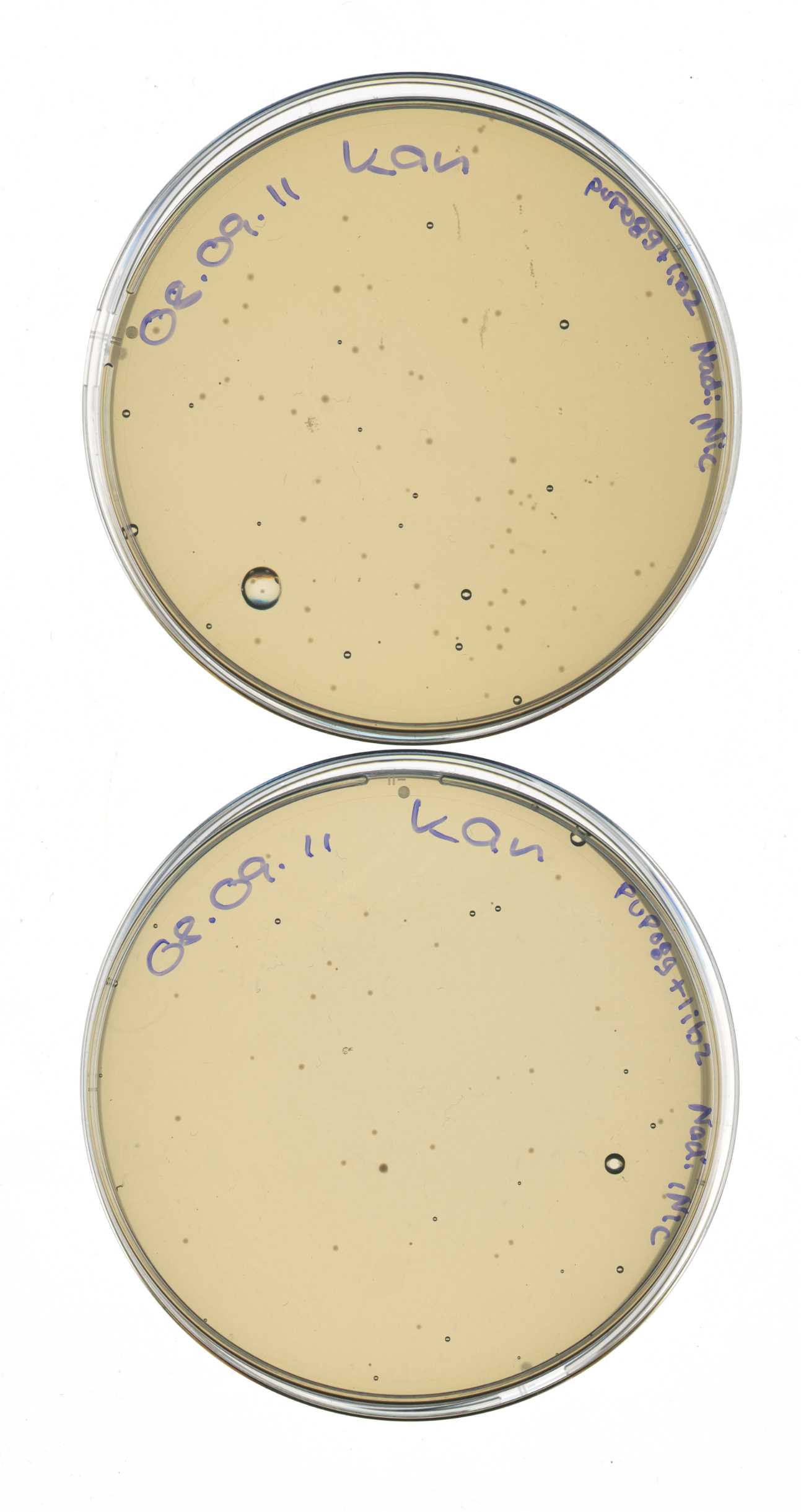
Glycerol stocks of pSB1C3_mdnBC and pUP089
Investigators: Nadine, Nicole, Niels
Aim: Preparing of glycerol stocks of pSB1C3_mdnBC and pUP089 for further use
Time: 2011-09-06, 8:30-9:15
Material:
Clones:
- pSB1C3_mdnBC, clone A, source, date
- pSB1C3_mdnBC, clone B, source, date
- pSB1C3_mdnBC, clone C
- pSB1C3_mdnBC, clone D
- pSB1C3_mdnBC, clone E
- pUP089 A clone A
- pUP089 A clone B
- pUP089 B clone A
- pUP089 B clone B
Glycerol
Method:
- adding of each 700 µl over night culture to 300 µl glycerol
- gently inverting
Results:
- glycerol stocks
Output:
stored in freezer (-21°C); glycerolstock box
| Number | Description | Strain | Resistance |
|---|---|---|---|
| G34 | pSB1C3_mdnBC clone A | XL1-blue | cm |
| G35 | pSB1C3_mdnBC clone B | XL1-blue | cm |
| G36 | pSB1C3_mdnBC clone C | XL1-blue | cm |
| G37 | pSB1C3_mdnBC clone D | XL1-blue | cm |
| G38 | pSB1C3_mdnBC clone E | XL1-blue | cm |
| G39 | pUP089 A clone A | XL1-blue | kana |
| G40 | pUP089 A clone B | XL1-blue | kana |
| G41 | pUP089 B clone A | XL1-blue | kana |
| G42 | pUP089 B clone B | XL1-blue | kana |
Miniprep of overnight cultures of pUP089 & psB1C3 + mdnBC
Investigators: Niels, Steffi
Materials:
- liquid culture of pSB1C3+ mdnBC & puP089
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| puP089 A a | 508 ng/µl |
| puP089 A b | 504 ng/µl |
| puP089 B a | 305 ng/µl |
| puP089 B b | 602 ng/µl |
| psB1C3 + mdnBC a | 444 ng/µl |
| psB1C3 + mdnBC b | 665 ng/µl |
| psB1C3 + mdnBC c | 661 ng/µl |
| psB1C3 + mdnBC d | 625 ng/µl |
| psB1C3 + mdnBC e | 609 ng/µl |
Further tasks:
- test digestion for confirmation
- sending for sequencing
check plates - resistance of competent E.coli ER2738 cells
Investigators: Niels, Katharina, Sandrina, Steffi
Aim: check resistance of competent cells – ER2738 (from 2011-09-05, Niels, Katharina, Sandrina, Steffi)
Materials:
- agar plates from competent cells - E.coli ER2738 from 2011-09-05 (San/ Ste)
Results:
- all competent cells - E.coli ER2738 grow on agar plates with Tetracycline and without antibiotics
- no E.coli ER2738 clones on agar plates with Ampicillin, Kanamycine, Chloramphenicol
Conclusions:
- competent cells - E.coli ER2738 work and can be used for transformation
PCR mdnABCDE w/ Long PCR Enzyme mix from Fermentas
Time: 2011-09-06
Investigators: Nicole, Nadine
Aim: generate BioBrick of mdnABCDE, amplify mdnABCDE for ligation in pSB1C3
Materials
- vector: pARW089 (8,6 ng/µl), pARW071 (6,3 ng/µl)
Materials:
- DNA polymerases: Long PCR Enzyme Mix (Fermentas)
- Template DNA: pARW071 (vector)
- dNTPs
- water
- Primer:
| # | Primer |
|---|---|
| 84 | pf_mdnABCDE89_EcoRI_NotI_XbaI (froward for pARW071 and pARW089) |
| 82 | r_mdnABCDE_iGEM (reverse for all) |
| 83 | pf_mdnABCDE89+T7_EcoRI_NotI_XbaI (froward for pARW089, generating mdnABCDE+T7) |
Protocol:
- mdnABCDE
- 2 µl vector
- 1 µl dNTPs (10 mM)
- 3 µl forward Primer
- 3 µl reverse Primer
- 0.3 µl Polymerase
- 5 µl Buffer Fermentas Long Enzyme Mix (+MgCl2)
- 35.3 µl water
- total volume: 50 µl
2. PCR program
- IGMDNBB for mdnB
- first steps: 10x
- second steps: 20x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 94°C | Hold |
| Initial denaturation | 94°C | 180 sec |
| Denaturation | 94°C | 15 s |
| Annealing | 59 | 30 s |
| Elongation | 68°C | 300 s |
| DenaturationII | 94°C | 15 s |
| AnnealingII | 72°C | 30 s |
| ElongationII | 72°C | 300 s + 2 s per each cycle s |
| Final Elongation | 68°C | 600 s |
Results:
- Output:
- mdnABCDE from pARW071, Nad/Nic, 6.9.11 (lane 2)
- mdnABCDE from pARW089, Nad/Nic, 6.9.11 (lane 3)
- mdnABCDE+T7 from pARW089, Nad/Nic, 6.9.11 (lane 4)
Further tasks:
- PCR purification
- digest
- ligation
PCR purification of mdnB
Investigators: Nadine, Nicole
Aim: Purification of mdnB (PCR product)
Time: 2011-09-06, 9:30-10:30
Material:
- PCR product of mdnB, PCR 2011-09-06, Nadja, Nicole, Jessica
- Machery-Nagel Nucleo Spin Extract II, PCR purification protocol
Method:
- done as described by manufacturer's protocol
- elution buffer
- elution volume 50 µl
Results:
- purified PCR product of mdnB
- cDNA = 44.6 ng/ µl
Output:
- stored in fridge, named: PCR product mdnB, pur., 2011-09-06
Further tasks:
- restriction enzyme digestion using SpeI and EcoRI
- Ligation in pSB1C3 (also digested with SpeI and EcoRI)
- Transformation in XL1-blue
Digest of purified PCR product mdnB and pSB1C3
Investigators: Niels
Materials:
- PCR product mdnB from 2011-9-5, Nad
- pSB1C3+mdnBC (a, b, c, d, e)
| backseat driver asks: why do you digest our almost finished biobricks (pSB1C3 carrying mdnBC)? Why don't you use the vector pSB1C3 from the plasmid box? |
- EcoRI, SpeI
- Buffer 4
- BSA
- H2O
Protocol:
- PCR product:
- 48 µl PCR product mdnB
- 1 µl EcoRI
- 1 µl SpeI
- 6 µl Buffer 4
- 0.6 µl BSA
- 3.4 µl H2O
- total volume: 60µl
- pSB1C3+mdnBC:
- 25 µl H2O and DNA (see below)
- 1 µl EcoRI
- 1 µl SpeI
- 3 µl Buffer 4
- 0.3 µl BSA
- total volume: 30µl
- pSB1C3+mdnBC dilutions
- a: 9 µl vector + 16 water
- b: 6 µl vector + 19 water
- c: 6 µl vector + 19 water
- d: 6.5 µl vector + 18.5 water
- e: ´6.5 µl vector + 18.5 water
- incubation 5 hrs at 37°C
Output:
- PCR mdnB, pur., dig., Nie, 6.9.11
- pSB1C3+mdnBC a, dig., Nie, 6.9.11
- pSB1C3+mdnBC b, dig., Nie, 6.9.11
- pSB1C3+mdnBC c, dig., Nie, 6.9.11
- pSB1C3+mdnBC d, dig., Nie, 6.9.11
- pSB1C3+mdnBC e, dig., Nie, 6.9.11
Further tasks:
- purification
- ligation
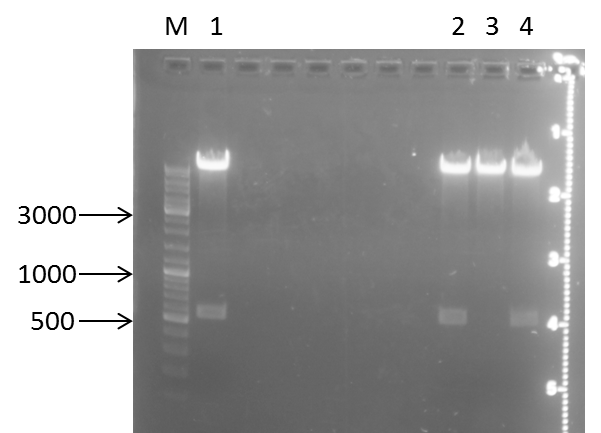
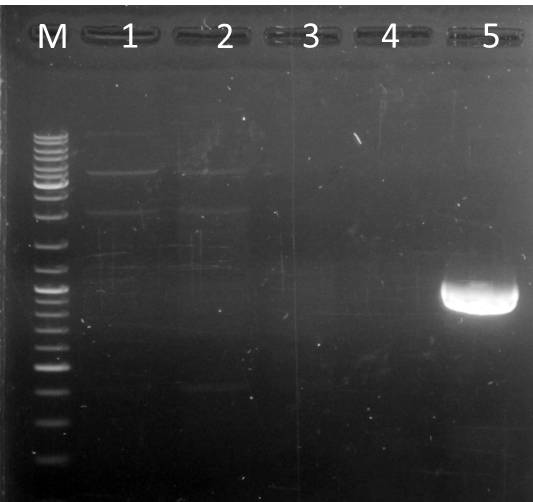
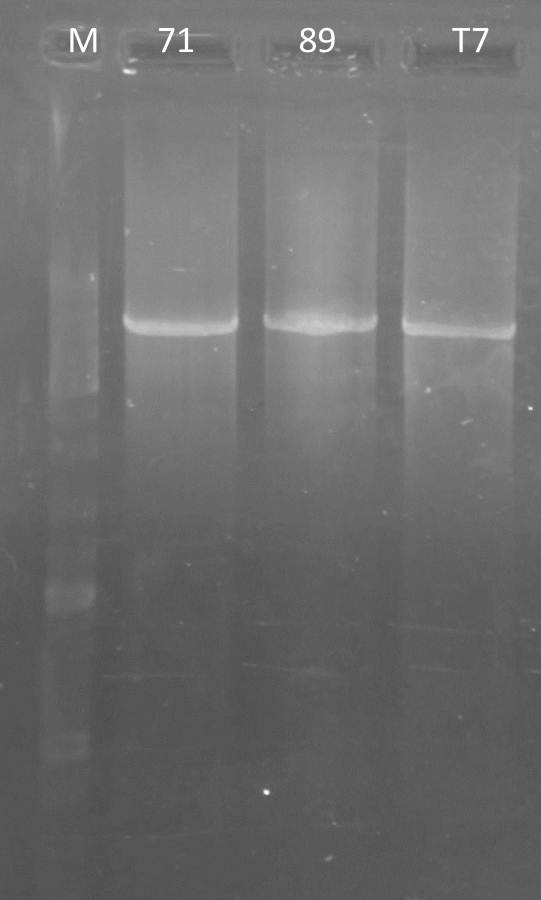
PCR: Agarose Gel: mdnB digest verification; pSBIC3_B digest verification and PCR mdnABCDE with Long Enzyme Mix verification
Time: 2011-09-06
Investigators:Nadja
Materials
- digested mdnB
- digested pSBIC3_B
- PCR outcome of mdnABCDE (89, T7, 71)
Production of one 0,8 % agarose gel
- 0,8 % gel: 0.4 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to the gel
Loading gels and running
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| M | Gene Ruler DNA Ladder Mix | 10µl (1:100) | |
| 1 | 71 | 2+ 3H2O+ 1 6x loading dye | 6600 |
| 2 | T7 | 2+ 3H2O+ 1 6x loading dye | 6600 |
| 3 | 89 | 2+ 3H2O+ 1 6x loading dye | 6600 |
| 4 | mdnB | 10+1 6x loading dye | 1031 |
| 5 | pSB1C3_B | 30+ 6µl loading dye | 2390, 243 |
Results:
Photos are already assigned to their appropriate experiments
Conclusions:
- test digest seems to be correct for mdnb (line 4), also the PCR was successful (line 1,2,3), only the pSB1C3_B digest did not worked out (line 5)
Further task:
- pSB1C3_A, C,D,E digest verification
PCR purification of PCR fragment mdnABCDE (T7, 89) and digested mdnB
Time: 2011-09-06
Investigators: Jessica, Nicole
Materials
- mdnABCDE+T7 from pARW089, Nad/Nic, 6.9.11
- mdnABCDE from pARW089, Nad/Nic, 6.9.11
- PCR mdnB, pur., dig., Nie, 6.9.11
- Macherey-Nagel Nucleo Spin Extract II kit
Protocol
- Protocol for PCR clean-up
- elution with 35 µl NE buffer (incubation for 2 min at 70°C before centrifuging)
Output:
- mdnABCDE+T7 from pARW089, pur., Nadja/Jessica, 6.9.11
- mdnABCDE from pARW089, pur., Nadja/Jessica, 6.9.11
- PCR mdnB, pur., dig., pur., Nadja, 6.9.11 (concentration: 65.7 ng/µl)
NOTE: mdnABCDE from pARW071, Nad/Nic, 6.9.11 and the library were mixed up, so PCR and fillin reaction were repeated
Further task:
- digest of mdnABCDE+T7 from pARW089, pur., Nadja/Jessica, 6.9.11 and mdnABCDE from pARW089, pur., Nadja/Jessica, 6.9.11 for ligation with pSB1C3
- ligation of PCR mdnB, pur., dig., pur., Nadja, 6.9.11 with pSB1C3
PCR: Agarose Gel: pSBIC3_A, C, D, E
Time: 2011-09-06
Investigators:Nadja
Materials
- digested pSBIC3_ACDE
| do you mean pSB1C3+mdnBC clone a to e? please use the same label names |
Production of one 0,8 % agarose gel
- 0,8 % gel: 0.4 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to the gel
Loading gels and running
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| M | Gene Ruler DNA Ladder Mix | 10µl (1:100) | |
| 1 | pSB1C3_A | 30+ 6x loading dye | 2390, 243 |
| 2 | pSB1C3_C | 30+ 6x loading dye | 2390, 243 |
| 3 | pSB1C3_D | 30+ 6x loading dye | 2390, 243 |
| 4 | pSB1C3_E | 30+ 6x loading dye | 2390, 243 |
Results:
Conclusions:
- worked out
Further task:
- take pSB1C3_E for gel extraction and ligate it with mdnB
Gel Extraction of pSB1C3_E
Time: 2011-09-06
Investigators: Jessica, Nadja
Materials
- pSBIC3_E
- use of Macherey Nagel Nucleo Spin Extrac II kit
Protocol
- 100mg gel – 200µl NT buffer ---- incubate for 5-10min at 50 C
- load sample on colum and spin for 1min at 11000 x g
- add 700µl NT3 and spin for 1min at 11000 x g
- spin again for 2 min at 11000 x g
- eluate DNA with 35µl NE by incubating 2 min at 50 C and spinning down for 1 min at 11000 x g
Results:
- measured concentration: 24,8 ng/ µl
Further task:
- ligation with mdnB
Ligation of pSB1C3_E and mdnB
Time: 2011-09-06
Investigators: Jessica, Nadja
Materials
- pSBIC3_E
- mdnB
- use Ligation Calculator
Protocol
Components
| Component | Molar Ratio | Concentration in ng/ µl | Length in bp | Max volume |
| Backbone | 1 | 24,8 | 2390 | 7 |
| Insert | 3 | 1031 | 65,7 | 7 |
Ligation mix
- 1µl Buffer
- 1µl Enzyme
- 2,6 µl Insert
- 5,4 µl Backbone
- 0 µl water
- total volume 10 µl
- at 14 C over night
Further task:
- Transformation
prepare samples for sequencing (created pARWIII)
Investigators: Sandrina, Sabine
Aim: sequencing to control ligation of geneIII into pARW089(GATC)
Material/Method:
- purified plasmids of clones 1, 2 and 4) (70 ng/µl, 20 µl total volume)
- primer mdnA_6 (10 pmol/µl, 30 µl total volume)
Further tasks: control sequence
Digestion of pSB1C3
Investigators: Sandrina, Sabine
Aim:
- cloning of biobricks mdnA, geneIII and mdna/geneIII (fusion gene) into pSB1C3
Materials/Methods:
- 8 µl pSB1C3 (ca 2 µg)
- 2 µl NEB 10x buffer 2
- 1 µl restriction enzyme XbaI
- 1 µ restriction enzyme PstI
- 0,2 µl BSA
- 7,8 µl water
- 3 h at 37°C
Further tasks:
- gel electrophoresis for purification
- ligation of mdnA, geneIII and mdna/geneIII (fusion gene) into pSB1C3
digestion of vector pARWIII
Investigators: Sandrina, Sabine
Aim: deletion of kanamycin gene in pARWIII (pARW089 containing geneIII
Materials/Methods:
- 2 µg sample (3 clones of pARWIII)
- 2 µl NEB 10x buffer 3
- 1 µl restriction enzyme NsiI
- add water: total volume 20 µl
- 3 h at 37°C
Further tasks:
- gel electrophoresis for purification
- re-ligation
Digestion of PCR products geneIII and mdnA for cloning into pSB1C3
Investigator: Sandrina, Sabine
Aim: cloning of mdnA, geneIII and mdnA/geneIII into pSB1C3
Material/Method:
- 30 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 4 µl NEB 10x buffer 2
- 0,4 µl BSA
- 3,6 µl water
- 1,5 h, 37°C
- 20 µl PCR product geneIII
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 3 µl NEB 10x buffer 2
- 4,7 µl water
- 0,3 µl BSA
- 1,5 h, 37°C
- 20 µl PCR product geneIII
- 1 µl restriction enzyme NgoMIV
- 1 µl restriction enzyme PstI
- 3 µl NEB 10x buffer 1
- 0,3 µl BSA
- 4,7 µl water
- 1,5 h, 37°C
- 20 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme AgeI
- 3 µl NEB 10x buffer 1
- 4,7 µl water
- 0,3 µl BSA
- 1,5 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation of mdnA and geneIII into pSB1C3
Agarose gel electrophoresis for purification of digested PCR products and vectors
Investigator: Sandrina, Sabine
Aim: control and purification of digested PCR products mdnA and geneIII
Material/Method:
- digested PCR products (mdnA and geneIII)
- digested vectors (pSB1C3 and pARWIII)
- 1 % agarose gel
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
- Gel Red
Ligation of geneIII and mdnA into pSB1C3
Investigator: Sabine
Aim: pSB1C3 containing mdnA, geneIII and mdnA/geneIII
Material/Method:
- 4,5 µl insert mdnA (7 ng/µ)
- 10 µl pSB1C3 (10 ng/µl)
- 2 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 2,5 µl water
- 1 h at room temperature
- 6,3 µl insert geneIII (10 µg/µl)
- 10 µl pSB1C3 (10 ng/µl)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 0,8 µl water
- 1 h at room temperature
- 3,2 µl insert mdnA (7,6 µ/µl)
- 6,5 µl insert geneIII (7 µg/µl)
- 7,3 µl pSB1C3 (10 ng/µl)
- 2 µl T4 ligase buffer (Fermentas)
- 1 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Re-ligation of NsiI-digested pARWIII
Investigator: Sabine
Aim: create pARW089 containing geneIII without kanamycin resistence (for library)
Material/Method:
- 17 µl NsiI-digested pARWIII (clones 1, 2 and 3)
- 1 µl T4 ligase buffer (Fermentas)
- 2 µl T4 Ligase (Fermentas)
- 1 h at room temperature
Further task: transformation
Transformation of created vectors in E. coli
Investigator: Sabine
Aim:amplification of vectors
Material:
- pARWIII without kanamycin resistence (clones 1, 2 and 3)
- pSB1C3 containing mdnA, geneIII or mdnA/geneIII
- agar plates containing chloramphenicol (pSB1C3)
- agar plates containing kanamycin, ampicillin(pPDV)
- agar plates containing kanamycin (pARW089)
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones
PCR of TorA-bla with clevage site of PreScission and TEV for BioBricks
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Stefan, Sebastian
Aim: get PCR fragment of TOR bla sequence for BioBricks
Methode:
Primer TEV:
(1)
(2)
Methode:
PCR
- Template: 1 µL (pJC354 <10ng)
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 5 µL 10 x Amplification buffer S
- 2 µL 25 mM MgCl2
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer)
- 35,5 µL of pure water
- 0,5 µL TaqPol
Program:
- Denat: 4 min 94°C
- 5x:
Denat: 1 min 94°C
Anneal: 1 min 51°C
Extend: 1 min 72°C
- 25x:
Denat: 1 min 94°C
Anneal: 1 min 61°C
Extend: 1 min 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification
PCR: mdnABCDE
Time: 2011-09-06
Investigators: Nicole, Nadja
Materials
- vector: pARW089 (2*)
- Long Enzyme Mix (+MgCl2) Buffer Fermentas
- Long Enzyme Polymerase Fermentas
- dNTPs (NEB)
- water
- Primer:
| # | Primer |
|---|---|
| x | pf_mdnABCDE089_NotI_SpeI_30.08. |
| x | r_mdnABCDE_iGEM |
Protocol:
- 2 µl pARW089 (diluted 1:100)
- 1 µl dNTPs (10 mM)
- 3 µl Primer forward (10 µM)
- 3 µl Primer backward (10 µM)
- 5 µl buffer
- 0.3 µl Ploymerase
- 35,7 µl water
- total volume: 50 µl
- mdnABCDE
2. PCR programs
- IGLONG2
Result
- PCR worked out
Cultures of pARW071 and pARW089 for HPLC
Time: 2011-09-06
Investigators: Nicole, Nadine, Jessica
Materials
- overnight cultures of pARW089 and pARW071
- 500mM Tris-HCl, pH 7.4
Protocol: (following protocol from Elke)
- inoculation of 400 ml LB medium with antibiotic
- at 37°C for about 7 h
- centrifuged 4x at 6000 g for 10 min in 50 ml falcons
- washed with Tris-HCl
Output:
- 4 falcons stored in -20°C (71 1, 71 2, 89 1, 89 2)
Overnight cultures of expression backbones
Time: 2011-09-06
Investigators: Jessica
Aim: Precultures for test of expression backbones
Materials
- plate of pSB1A3_Ara_YFP (from 2011-09-03)
- test cultures of pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP (from 2011-09-06)
- LB medium, Kan, Amp
Method:
- inoculation of 10 ml LB medium with antibiotic
- overnight at 37°C
Further tasks:
- testing expression
88th Labday 2011-09-07
Digestion and Ligation of TorA-bla with cleavage site of PreScission and TEV for BioBricks and TEV / PreScission biobricks
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul
Aim: purification and digestion PCR fragment (produced: 06.09.2011) of TOR bla sequence for BioBricks
Methode:
PCR-Products were purificated using Nucleospin Extract II KIT.
- The torA-bla construct was digested with AgeI and XbaI
- The vector (BBa_K404304_pSB1C3) for torA-bla was digested with AgeI and XbaI
- PreScission and Tev proteases were digested with EcoRI and SpeI
- The vector (BBa_K404304_pSB1C3) for proteases was digested with EcoRI and SpeI
The digestion was allowed to proceed for 2h at 37°C.
- Digested products were resolved on 1% agarose gel, the corresponding bands were excissed and extracted from the gel using Nucleospin gel extraction KIT
- Expected bands:
- Tev: 760 bp (bands: Tev=2,Tev2=3,Tev4=4, Tev7=5)
- Pre: 570 bp (bands: Pre1=15, Pre2=16)
- TorA-bla: 1001 bp (bands: Cs_TevI=6, Cs_TevII=7, Cs_TevIII=8, Cs_PreI=10, Cs_PreII=11)
- Vector: ~2632 bp (bands: Vector for proteases=14, vector for Tor_bla=17)
- The excised fragments were ligated into corresponding digested and excised vectors: Tev2, Tev3, Tev4, Tev7 and Pre1 and Pre2 into vector for proteases; the torA-bla fragments were ligated into vector for TorA-bla)
- ligation was calculated with gibthon ligation calculator ([http://www.gibthon.org/ligate.html Klick]) with a molar ration of 1:3 of plasmid to insert.
Proteases:
- 1 µl T4-ligation (Fermentas)
- 1 µl Ligase Buffer
1.4 µl vector - 6.6 µl Tev
1.2 µl vector - 6.8 µl Tev2
0.7 µl vector - 7.3 µl Tev4
1.5 µl vector - 6.5 µl Tev7
0.3 µl vector - 7.7 µl Pre1
0.4 µl vector - 7.6 µl Pre2
- = 6x10 µl ligation batches
TorA-bla
- 1 µl T4-ligation (Fermentas)
- 1 µl Ligase Buffer
2.2 µl vector - 5.8 µl Cs_PreI
2.4 µl vector - 2.6 µl Cs_PreII
1.5 µl vector - 6.5 µl Cs_TevI
1.7 µl vector - 6.3 µl Cs_TevII
2.1 µl vector - 5.9 µl Cs_TevIII
- = 5x10 µl ligation batches
- ligation was allowed to proceed for 1h at room temperature.
- 2µl of each ligation batch was transformed into competent Xl1-blue cells.
- Cells were plated in Cm containing agar plates
Oligo-Fillin for mdnA-Library (repetition)2
Time: 2011-09-07, 7:20-8:40
Investigators: Nadine, Nicole
Aim: Production of a library carrying mutated mdnA ß-lactamase-screening (carrying AatII restriction site)
Material:
- oligos (25 mM, HPLC purified, ordered by Sigma Aldrich):
- o_mdnA_library, 76
- o_focused_library_2, 74
- 10x Klenow buffer
- dNTPs (10 mM)
- H2O
Method:
- Reaction mix (total volume: 50 µl)
- 3 µl o_mdnA-library, 76
- 3 µl o_focused_library_2, 74
- 5 µl dNTPs
- 5 µl Klenow buffer
- 34 µl H2O
- Reaction conditions
- Program: ORIGAMI1
- after finishing program ORIGAMI1:
- addition of 1 µl Klenow-fragment
- incubation 1 hour, 37°C
Results:
- hybridized oligo for production of mdnA-library
Output:
- hybridization product, named foc 2
- stored in thermocycler
Further tasks:
- PCR purification
- restriction enzyme digestion using SfoI and AatII, also of pUP089
Preparing samples mdnC, mdnD, mdnE and mdnABC for sequencing
Time: 2011-09-07,
Investigators: Katharina, Nadine, Nicole
Purification of Fillin mdnA-Lib2
Time: 2011-9-7,
Investigators: Nadine, Nicole
Aim: Purification of oligos for focused library 2
Materials
- product of filled-in reaction, 2011-09-07, Nadine, Nicole
- Machery-Nagel Nucleo Spin Extract II kit
Method:
- application based on manufacturer’s protocol for PCR product purification
- exceptions for elution procedure:
- H2O used for elution
- elution volume V=25 µl
- before centrifugation (for 1 min) incubation 5 min at 70°C
- DNA concentration measured by Nanotrop
Results:
- cDNA: 113.3 ng/ µl
Output:
- purified oligos (focused library 2)
Further tasks:
- digestion using SfoI and AatII
- digestion of pUP089 using SfoI and AatII
- Purification of both
- Ligation of pUP089 and oligos (focused library 2)
digest of purified Fillin mdnA-Lib2 and pUP089
Time: 2011-9-7,
Investigators: Nadine, Nicole
purification of digested pSB1C3 and pUP089
Time: 2011-09-07,
Investigators: Niels, Jessica, Katharina
Ligation of Fillin mdnA-Lib2 and pUP089
Time: 2011-9-7,
Investigators: Nadja
Materials
- product of digested and purified Fillin mdnA Lib2 (2011-9-7, Nicole, Nadine) = 123,0 ng/ µl
- pUP089 = 14,9 ng/ µl
- Quick Ligase Buffer
Protocol:total volume of 20µl+
| backseat driver asks: where is the ligation control? we talked about this several times!!!!!!!!! now we have to repeat this -.- and time designation would be also nice |
- 10 µl Quick ligase buffer
- 1µl Enzyme
- 1µl Insert
- 8µl backbone
- at 16 °C in PCR machine
Further tasks:
- transformation
Ligation of mdnABCDE in pSB1C3
Time: 2011-9-7,
Digest of purified PCR product mdnABCDE and pSB1C3
Time: 2011-9-7, 8:30-12:15
Investigators: Nadine, Nicole
Aim: generate a BioBrick of mdnABCDE, digest for ligation in pSB1C3
Materials:
- PCR products:
- mdnABCDE from pARW089 from 2011-9-6, Nad/Nic
- mdnABCDE+T7 pARW089 from 2011-9-6, Nad/Nic
- mdnABCDE from pARW089 or pARW071 from 2011-9-6, Nadj
- BBa_K404304_pSB1C3 (#3), 284.6 ng/µl
- EcoRI, SpeI
- Buffer 4
- BSA
- H2O
Protocol:
- PCR product:
- 30 µl PCR product
- 1 µl EcoRI
- 1 µl SpeI
- 5 µl Buffer 4
- 0.6 µl BSA
- 12.5 µl H2O
- total volume: 50µl
- vector:
- 5 µl vector
- 1 µl EcoRI
- 1 µl SpeI
- 2 µl Buffer 4
- 0.2 µl BSA
- 10.8 µl H2O
- total volume: 20µl
- incubation 3 hrs at 37°C (start: 9:15)
Output:
- mdnABCDE from pARW089, Eco, Spe, from 2011-9-7, Nad/Nic
- mdnABCDE+T7 pARW089, Eco, Spe from 2011-9-7, Nad/Nic
- mdnABCDE from pARW089, Eco, Spe or pARW071 from 2011-9-7, Nad/Nic
- BBa_K404304_pSB1C3, Eco, Spe, from 2011-9-7, Nad/Nic
Further tasks:
- agarose gel
- purification
- ligation
test digest of mdnBC
Time: 2011-9-7,10:30-
Investigators: Nadine, Nicole
Aim: prove of Insert (mdnBC)
Materials:
- mdnBC klon a-e, Nie, 6.9.11
- HindIII, AvaI, (NEB)
- Buffer 2 (NEB, 10x)
- water
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnBC | 4419 | HindIII, AvaI | 2 | 363, 836, 3220 |
Digestion protocol
- 1 µl DNA
- 0.5 µl HinIII
- 0.5 µl AvaI
- 2 µl 10x buffer
- 16 µl pure water
- total: 20µl
- 37°C for 2 hrs
Conclusions:
Further tasks:
- agarose gel
- sequencing
test pSB1A3_Ara_YFP and pSB1A3_Lac_YFP (expression backbones)
Time: 2011-9-7,12
Investigators: Nadine, Nicole, Jessica
Method:
- pSB1A3_Lac_YFP:
- ODs at 16:45: 0.095 (1) and 0.082 (2, control)
- no induction
- pSB1A3_Ara_YFP:
- ODs at 17:30 : 0.657 (1) and 0.713 (2, control)
- induction with arabinose, end concentration 5 mM
Results:
- fluorescence didn't increase, curve flattened
PCR: mdnABCDE
Time: 2011-09-06
Investigators: Nadja
Materials
- vector: pARW071 (1)
- Long Enzyme Mix (+MgCl2) Buffer Fermentas
- Long Enzyme Mix Polymerase Fermentas
- dNTPs (NEB)
- water
- Primer:
| # | Primer |
|---|---|
| 84 | pf_mdnABCDE089_NotI_SpeI_30.08. |
| 82 | r_mdnABCDE_iGEM |
Protocol:
- 2,6 µl pARW08 (diluted 1:100)
- 1 µl dNTPs (10 mM)
- 3 µl Primer forward (10 µM)
- 3 µl Primer backward (10 µM)
- 5 µl buffer
- 0.3 µl Ploymerase
- 35,4 µl water
- total volume: 50 µl
- mdnABCDE
2. PCR programs
- IGLONG2
Further task
- gel verification
- purification
Overnight cultures of expression backbones and pUP089
Time: 2011-09-07, 18:00
Investigators: Niels
Aim: Precultures for test of expression backbones and glycerol stocks
Materials
- plates of pSB1A3_Ara_YFP, pSB1A3_Lac_YFP, pSB1K3_Ara_CFP, pSB1K3_Lac_CFP (from 2011-09-03)
- plates of pUP089 A and B
- LB medium, Kan, Amp
Method:
- inoculation of 10/5 ml LB medium with antibiotic
- overnight at 37°C
Further tasks:
- testing expression
- glycerol stocks !!!!!!!
Repeated test digest of mdnBC
Time: 2011-9-7, 20:00
Investigators: Jessica, Niels, Nadja
Aim: prove of Insert (mdnBC)
Materials:
- mdnBC clone b, Nie, 6.9.11
- HindIII, AvaI, PstI-HF, XmaI (NEB)
- Buffer 2 and 4 (NEB, 10x)
- water
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnBC | 4419 | HindIII, AvaI | 2 | 363, 836, 3220 |
| mdnBC | 4419 | PstI-HF, XmaI | 4 | 1559, 2868 |
Digestion protocol
- 1 µl DNA
- 0.5 µl per enzyme
- 2 µl 10x buffer
- 16 µl pure water
- total: 20µl
- 37°C for overnight (on thermomixer in iGEM lab)
Agarose gel:
Conclusions:
Further tasks:
- sequencing
Production of phages containing pPDV089 in ER2738 cells
Investigators: Sabine, Sandrina, Steffi
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- first step: amplification of cells containing pPDV089 (clone: 2S14):
- 50 ml DYT medium will be inoculated with the cells, so that OD600 = 0.1
- add antibiotics tetracyclin and ampicillin to the medium
- cells should be incubated 37 °C till OD600 = 0.3-0.5 (here: 0.356) is reached
- second step: infection with helper phages
- add helper phages 10^11 phages/50 ml (3,5 µl)
- incubate for 10 min at 37°C (without shaking!)
- add 0,5 mM IPTG
- incubate 50 min at 28°C and rpm
- add 70 µg/ml kanamycin and incubate for 5 h at 28°C (...)
- third step: phage purification
- centrifuge cell culture at 5000 x g/ 15 min
- fill supernatant in a new 50 ml falcon and centrifuge again (5000 g/15 min)
- 40 ml of the supernatant with 8 ml PEG-NaCl (20% (w/v) PEG-8000, 2,5 M NaCl)
- incubate over night at 4°C
Further tasks:
- go on with phage purification
SDS- PAGE for Western Blotting and coomassie staining of purified phages
Investigators: Sabine, Sandrina, Sebastian
Aim: control if geneIII-mdnA is expressed on the phages produced in XL1-blue cells
Method/Materials:
Further tasks:
- Western Blot and coomassie staining
Western Blot
Investigators: Sabine, Sandrina
Aim: control if geneIII-mdnA is expressed on the phages produced in XL1-blue cells
Method/Materials:
- membrane impregnated in methanol
- whatman paper
- blotting buffer
- blotting chamber
- 1 h at 200 mA
- blocking over night in TBS with 5% milk powder
Further tasks:
- antibody detection
Coomassie staining
Investigators: Sabine, Sandrina
Aim: control if geneIII-mdnA is expressed on the phages produced in XL1-blue cells
Method/Materials:
- 20 ml water
- 20 ml methanol
- 60 ml coomassie stock solution
- gel storage over night in coomassie solution shaking
Further tasks:
- destaining of the gel
overnight culture of picked E. coli clones transformed with pARWIII (pARW089 containing geneIII) to control deletion of kanamycin gene, pSB1C3 containing mdnA, geneIII or mdnA/geneIII
Investigators: Sandrina, Sabine
Aim: control deletion of kanamycin gene in created pARWIII, control ligation of mdnA, geneIII and mdnA/geneIII into pSB1C3
Method/Materials:
- 5 clones from pARW089 with geneIII (kanamycin)
- 5 clones from pSB1C3 with mdnA (chloramphenicol)
- 5 clones from pSB1C3 with geneIII (chloramphenicol)
- 5 clones from pSB1C3 with mdnA/geneIII (chloramphenicol)
- 5 ml LB medium per clone
- storage over night at 37°C and 800 rpm
Further tasks:
- test digestion
89th Labday 2011-09-08
Oligo-Fillin for mdnA-library 2 (repetition) and for Phage-display-mdnA-library (carrying AgeI restriction site)
Investigators: Nicole, Nadine
Time: 2011-09-08, 11:00-12:30
Aim: Production of libraries carrying mutated mdnA, one for phage-display (carrying ageI restriction site) and one for ß-lactamase-screening (carrying AatII restriction site)
Material:
- oligos (25 mM, HPLC purified, ordered by Sigma Aldrich):
- o_mdnA_library, 76 à for both libraries
- o_focused_library_2, 74 à for AatII carrying library
- o_focused_library_AgeI, à for AgeI carrying library
- 10x Klenow buffer
- dNTPs (10 mM)
- H2O
Method:
- Reaction mix (total volume: 50 µl)
- 3 µl fw-oligonucleotide (o_mdnA-library, 76)
- 3 µl rev-oligonucleotide (either o_focused_library_2, 74 or o_focused_library_AgeI, )
- 5 µl dNTPs
- 5 µl Klenow buffer
- 34 µl H2O
- Reaction conditions
- Program: ORIGAMI1
- after finishing program ORIGAMI1:
- addition of 1 µl Klenow-fragment
- incubation 1 hour, 37°C
Results:
- hybridized oligos for production of mdnA-library
Output:
- two hybridization products
- 1) AgeI-library, named AgeI
- 2) AatII-library, named foc 2
- stored in thermocycler
Further tasks:
- PCR purification
- restriction enzyme digestion using SfoI and either AatII or AgeI, also of either pUP089 or pARWIII
Transformation of XL1 with different constructs
Time: 2011-09-08, 12:30-15:00
Investigators: Nadine, Nicole
Materials:
- XL1-Blue
- ligation products:
- pSB1C3+mdnABCDE+T7, Niels, 7.9.11, resistance: cm
- pSB1C3+mdnABCDE from pARW089, Niels, 7.9.11, resistance: cm
- pSB1C3+mdnB, Jes/Nadja, 6.9.11, resistance: cm
- pSB1C3+H20 (control), Jes/Nadja, 6.9.11, resistance: cm
- pUP089+lib2, Nadja, 7.9.11, resistance: kan
- plasmids from miniprep (Kat, 2011-8-20):
- pSB1A3_Ara_YFP clone A (#1), resistance: amp
- pSB1A3_Lac_YFP clone B (#4), resistance: amp
- pSB1K3_Ara_CFP clone A (#25), resistance: kan
- pSB1K3_Lac_CFP clone B (#21), resistance: kan
- pSB1K3_Lac_YFP clone C (#18), resistance: kan
- pSB1K3_Lac_YFP clone C (#18), resistance: kan
- antibiotics from stock:
- kanamycin
- ampiciline
- chloramphenicol
- LB-Agar
Method:
- ligation products:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 30 min on ice,
- heat shock 60 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (20 ml Agar and 20 µl antibiotic (see above) per plate)
- storage over night at 37°C (start: 15:00)
Further tasks:
- Picking clones for overnight culture (except for library 2)
- Producing glycerol stocks
- count colonies of pUP089_lib2 and collect them w/ LB
Output:
- Plates w/ XL1 blue:
- pSB1C3+mdnABCDE+T7, Niels, 7.9.11, resistance: cm
- pSB1C3+mdnABCDE from pARW089, Niels, 7.9.11, resistance: cm
- pSB1C3+mdnB, Jes/Nadja, 6.9.11, resistance: cm
- pSB1C3+H20 (control), Jes/Nadja, 6.9.11, resistance: cm
- 10x pUP089+lib2, Nadja, 7.9.11, resistance: kan
- pSB1A3_Ara_YFP clone A (#1), resistance: amp
- pSB1A3_Lac_YFP clone B (#4), resistance: amp
- pSB1K3_Ara_CFP clone A (#25), resistance: kan
- pSB1K3_Lac_CFP clone B (#21), resistance: kan
- pSB1K3_Lac_YFP clone C (#18), resistance: kan
- Ligation products stored in freezer, green box, mdn-BioBricks
- pSB1C3+mdnABCDE+T7, pARW089, 7.9.11
- pSB1C3+mdnABCDE, pARW089, 7.9.11
- pSB1C3+mdnB, Nadja/Jes, 6.9.11
- pSB1C3+H20, Nadja/Jes, 6.9.11
- Expresion backbones stored in white/colorless box w/ label: Expression Backbones 2
Purification of Fillin mdnA-Library_2 (carrying either AgeI or AatII restriction site) and PCR product of mdnABCDE from pARW071
Time: 2011-9-8,
Investigators: Nadine, Nicole
Aim: Purification of oligos for focused library 2 for phage display and ß-lactamase screening and purification of PCR product mdnABCDE from pARW071 for further use
Materials
- product of filled-in reaction, 2011-09-08, Nadine, Nicole
- Age (mdnA library for phage display)
- foc 2 (mdnA library for ß-lactamase screening)
- PCR product of mdnABCDE from pARW071 (named mdnA71), 2011-09-07, Nadja
- Machery-Nagel Nucleo Spin Extract II kit
Method:
- application based on manufacturer’s protocol for PCR product purification
- exceptions for elution procedure:
- H2O used for elution
- elution volume V=25 µl
- before centrifugation (for 1 min) incubation 5 min at 70°C
- DNA concentration measured by Nanotrop
Output:
- purified oligos
- mdnA focused library 2 (carrying AatII restriction site), named Age, stored in freezer
- mdnA focused library 2 (carrying AgeI restriction site), named foc 2, stored in freezer
- mdnABCDE from pARW071, named mdnA71, stored in freezer
Further tasks:
- digestion of foc 2 using SfoI and AatII
- digestion of pUP089 using SfoI and AatII
- digestion of Age using SfoI and AgeI
- digestion of pARWIII using SfoI and AgeI
- digestion of mdnA71 using EcoRI and SpeI
- digestion of pSB1C3 using EcoRI and SpeI
Restriction enzyme digestion of mdnA focused library 2 carrying AatII restriction site and pUP089
Investigators: Nicole, Nadine
Time: 2011-09-08,
Aim: Digest of hybridized oligos for further production of mdnA focused library
Material:
- clones
- purified mdnA oligos carrying AatII restriction site, named foc 2, 2011-09-08, Nicole/Nadine
- pUP089, clone A I, Nadine
- NEB buffer 4
- AatII
- SfoI
Method:
- Mix for digestion of foc 2
- 23 µl PCR purification product foc 2 (mdnA oligos)
- 2 µl AatII
- 2 µl SfoI
- 3 µl NEB buffer 4
- Mix for digestion of pUP089
- 4 µl DNA pUP089
- 2 µl AatII
- 2 µl SfoI
- 2 µl NEB buffer 4
- 10 µl H2O
- Incubation for 2 hours, 37 °C
Results:
- digested oligos for library
- digested pUP089
Output:
- digested oligos for mdnA library (carrying AatII restriction site), named foc 2, stored in ice box
- digested pUP089, named pUP089, stored in ice box
Further tasks:
- PCR purification (foc 2) resp. agrose gel and gel extraction (pUP089)
- ligation of both
Restriction enzyme digestion of mdnABCDE71 and pSB1C3
Investigators: Nicole, Nadine
Time: 2011-09-08,
Aim: Digest of PCR product mdnABCDE from pARW071 and pSB1C3 to prepare a biobrick
Material:
- clones
- purified PCR product mdnABCDE, named mdnABCDE71, 2011-09-08, Nicole/Nadine
- pSB1C3,
- NEB buffer 4
- EcoRI-HF
- SpeI
- BSA
Method:
- Mix for digestion of mdnABCDE from pARW071 (total volume 50 µl)
- 23 µl PCR purification product foc 2 (mdnA oligos)
- 1 µl EcoRI-HF
- 1 µl SpeI
- 5 µl NEB buffer 4
- 0.6 µl BSA
- 19.5 H2O
- Mix for digestion of pSB1C3 (total volume 20 µl)
- 5.8 µl DNA pSB1C3
- 1 µl EcoRI-HF
- 1 µl SpeI
- 2 µl NEB buffer 4
- 10 µl H2O
- 0.2 µl BSA
- Incubation for 2 hours, 37 °C
Results:
- digested oligos for library
- digested pARWIII
Output:
- digested mdnABCDE from pARW071, named digested mdnABCDE71, stored in ice box
- digested pSB1C3, named digested pSB1C3, stored in ice box
Further tasks:
- PCR purification (mdnABCDE71) resp. agrose gel and gel extraction (pSB1C3)
- ligation of both
Restriction enzyme digestion of mdnA focused library 2 carrying AgeI restriction site and pARWIII
Investigators: Nicole, Nadine
Time: 2011-09-08,
Aim: Digest of hybridized oligos for further production of mdnA focused library for phage-display
Material:
- clones
- purified mdnA oligos carrying AgeI restriction site, named Age, 2011-09-08, Nicole/Nadine
- pARWIII, clone 1_1, Sandrina, 2011-09-08
- NEB buffer 4
- AgeI
- SfoI
Method:
- Mix for digestion of Age
- 22.5 µl PCR purification product foc 2 (mdnA oligos)
- 2.5 µl AgeI
- 2 µl SfoI
- 3 µl NEB buffer 4
- Mix for digestion of pARWIII
- 6.5 µl DNA pARWIII
- 2.5 µl AgeI
- 2 µl SfoI
- 3 µl NEB buffer 4
- 6 µl H2O
- Incubation for 2 hours, 37 °C
Results:
- digested oligos for library
- digested pARWIII
Output:
- digested oligos for mdnA library (carrying AgeI restriction site), named Age, stored in ice box
- digested pARWIII, named digested-pARWIII, stored in ice box
Further tasks:
- PCR purification (Age) resp. agrose gel and gel extraction (pARWIII)
- ligation of both
PCR of TorA-bla with clevage site of PreScission and TEV for BioBricks
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Stefan, Sebastian
Aim: get PCR fragment of TOR bla sequence for BioBricks
Methode:
Primer TEV:
(1)
(2)
Methode:
PCR
- Template: 1 µL (pJC354 <10ng)
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 10 µL 5 x Amplification buffer
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer)
- 35 µL of pure water
- 0,5 µL Fusion
Program:
- Denat: 4 min 94°C
- 30x:
Denat: 30 sec 94°C
Anneal: 30 sec 72°C
Extend: 1 min 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification
Test digest of pSB1C3+mdnDE
Time: 2011-9-08, 17:30-20:30
Investigators: Jessica
Aim: prove of Insert (mdnDE)
Materials:
- pSB1C3+mdnBC clones K1-K6, Kat, 8.9.11
- concentrations:
| Sample | Concentration in ng/µl |
| clone K1 | 835.1 |
| clone K2 | 775.6 |
| clone K3 | 488.1 |
| clone K4 | 639.3 |
| clone K5 | 554.3 |
| clone K6 | 551.5 |
- HpaI (NEB)
- Buffer 4 (NEB, 10x)
- water
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnDE | 5154 | HpaI | 4 | 1329, 3825 |
Digestion protocol
- 1 µl DNA
- 0.5 µl HpaI
- 2 µl 10x buffer
- 16 µl pure water
- total: 20µl
- 37°C for
Agarose gel
| lane | Sample | Volume in µl |
| M | Gene Ruler DNA Ladder Mix (1:10) | 20µl |
| 1 | - | |
| 2 | clone K1 | 20 + 4x loading dye |
| 3 | clone K2 | 20 + 4x loading dye |
| 4 | clone K3 | 20 + 4x loading dye |
| 5 | clone K4 | 20 + 4x loading dye |
| 6 | clone K5 | 20 + 4x loading dye |
| 7 | clone K6 | 20 + 4x loading dye |
Conclusions:
- clones K1, K2 and K5 are ok
Further tasks:
- sequencing
Overnight cultures of pSB1C3+myc
Time: 2011-09-08, 18:30
Investigators: Jessica
Materials
- plates of pSB1C3+mycN and pSB1C3+mycC
- LB medium, Cm
Method:
- inoculation of 5 ml LB medium with Cm
- overnight at 37°C
Further tasks:
- test digest
- sequencing
Sidedirected mutagenesis of TEV protease and PreSciccion protease
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim: Sidedirected mutagenesis of TEV and PreSciccion protease
Materials:
- TEV protease vector (P9 by Gunther Stier)
- PreSciccion protease vector (P10 by Gunther Stier)
- different Primers
- Phusion polymerase kit by NEB
Methode:
PCR
- Template: 1 µL (pJC354 <10ng)
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 10 µL 5 x Phusion HF Buffer
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer, see list below)
- 32,5 µL of pure water
- 0,5 µL Phusion HF polymerase
Program:
- Denat: 30 sec 98°C
- 30x
Denat: 10 sec 98°C
Anneal: 15 sec 69°C (with gradient of +/- 4°C)
Extend: 15 sec 72°C
- Final Extend: 10min 72°C
Used Primers:
Fragment 1 of TEV protease (T1):
- f_TEV_AraFusion_NgoMIV
- r_TEV_ACCAGC
Fragment 2 of TEV protease (T2):
- f_TEV_ACCAGC
- r_TEV_iGEM_BamHI
Fragment 1 of PreSciccion protease (P1):
- f_Pre_AraFusion_NgoMIV
- r_Pre_ACCAGC
Fragment 2 of PreSciccion protease (P2):
- f_Pre_ACCAGC
- r_Pre_tm_Xba208_A-T
Fragment 3 of PreSciccion protease (P3):
- f_Pre_tm_Xba208_A-T
- r_Pre_tm_Xba280_A-T
Fragment 4 of PreSciccion protease (P4):
- f_Pre_tm_Xba280_A-T
- r_Pre_iGEM_BamHI
Further Tasks:
PCR purification, analytical Gel to check sizes of the amplified fragments, assembly PCR of Fragment
Analytical gelelctrophoresis and PCR clean up of sidedirected mutated fragments T1, T2, P1, P2, P3 and P4 http://syntbio.net/iGEM/wiki2011/index.php/Labjournal_september#Sidedirected_mutagenesis_of_TEV_protease_and_PreSciccion_protease
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim: Check the size of amplified fragments and clean them up after PCR
Materials:
- PCR fragments P1 to P4 and T1 and T2
- Fermentas 6x loading dye
- 100bp ladder (Gene Ruler by Fermentas)
- Macherey-Nagel Nucleo Spin II Kit for clean up
Methode:
- Clean up after manual for PCR purification of the used Kit
- gel electophoresis (2% agarose gel with analytical gel pockets): 30 min at 110 V
Results:
The sizes of the fragments of the PCR products are matching the fragment sizes extracted with Geneious:
- T1
- sizes in Geneious: 411 bp
- estimated sizes in gel: 410 bp
- T2
- sizes in Geneious: 389 bp
- estimated sizes in gel: 400 bp
- P1
- sizes in Geneious: 167 bp
- estimated sizes in gel: 170 bp
- P2
- sizes in Geneious: 96 bp
- estimated sizes in gel: 100 bp
- P3
- sizes in Geneious: 99 bp
- estimated sizes in gel: 100 bp
- P4
- sizes in Geneious: 311 bp
- estimated sizes in gel 310 bp
File:UP AG 2011-09-08-0140 muta TEV-Pre.JPG
DNA-Fragments after sidedirected mutagenesis:
- TEV fragment 1 (T1) c=
- TEV fragment 2 (T2) c=
- PreSciccion fragment 1 (P1) c=
- PreSciccion fragment 2 (P2) c=
- PreSciccion fragment 3 (P3) c=
- PreSciccion fragment 4 (P4) c=
Output:
Fragments T1 and T2, P1-P4 with expected sizes.
Further Tasks:
Merge of PCR fragments P1 and P2 into P5, P3 and P4 into P6, T1 and T2 into TEV-protease.
Assembly PCR of mutated TEV protease and PreSciccion protease fragments
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Stefan, Sebastian
Aim: Assembly of mutated TEV and PreSciccion protease fragments
Materials:
- TEV protease fragments T1 and T2 --> TEV protease (total mutated)
- PreSciccion protease fragments P1 and P2 --> P5
- PreSciccion protease fragments P3 and P4 --> P6
- different Primers
- Phusion polymerase kit by NEB
Methode:
PCR
- Template: 2 µL (1µl of each fragment after sizes were checked and fragments got cleaned up with PCR clean up kit from Macherey-Nagel (Nucleo Spin Kit))
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 10 µL 5 x Phusion HF Buffer
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer, see list below)
- 31,5 µL of pure water
- 0,5 µL Phusion HF polymerase
Program:
- Denat: 30 sec 98°C
- 30x
Denat: 10 sec 98°C
Anneal: 15 sec 70°C (with gradient of +/- 2°C)
Extend: 20 sec 72°C
- Final Extend: 10min 72°C
Used Primers:
Assembly of fragment 1 and fragment 2 of TEV protease (T1+T2) --> mutated TEV protease:
- f_TEV_AraFusion_NgoMIV
- r_TEV_iGEM_BamHI
Assembly of fragment 1 and fragment 2 of PreSciccion protease (P1+P2)--> mutated PreSciccion portease fragment 5 (P5):
- f_Pre_AraFusion_NgoMIV
- r_Pre_tm_Xba208_A-T
Assembly of fragment 3 and fragment 4 of PreSciccion protease (P3+P4)--> mutated PreSciccion portease fragment 6 (P6):
- f_Pre_tm_Xba208_A-T
- r_Pre_iGEM_BamHI
Further Tasks:
PCR purification, analytical Gel to check sizes of the amplified fragments, assembly PCR of Fragment
Analytical gelelctrophoresis and PCR clean up of sidedirected mutated fragments TEV protease, P5 and P6 http://syntbio.net/iGEM/wiki2011/index.php/Labjournal_september#Assembly_PCR_of_mutated_TEV_protease_and_PreSciccion_protease_fragments
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim: Check the size of amplified fragments and clean them up after PCR
Materials:
- PCR fragments P5, P6 and TEV protease
- Fermentas 6x loading dye
- 100bp ladder (Gene Ruler by Fermentas)
- Macherey-Nagel Nucleo Spin II Kit for clean up
Methode:
- Clean up after manual for PCR purification of the used Kit
- gel electophoresis (1.5 % agarose gel with analytical gel pockets): 30 min at 110 V
Results:
The sizes of the fragments of the PCR products are matching the fragment sizes extracted with Geneious:
- TEV
- sizes in Geneious:
- estimated sizes in gel
- P5
- sizes in Geneious:
- estimated sizes in gel
- P6
- sizes in Geneious:
- estimated sizes in gel
(Picture from GelDoc and sizes will be added as soon as possible)
DNA-Fragments after sidedirected mutagenesis:
- TEV protease c=
- PreSciccion fragment 5 (P5) c=
- PreSciccion fragment 6 (P6) c=
Further Tasks: Merge of PCR fragments P5 and P6 into PreSciccion protease, digestion of TEV protease with NgoMIV and BamHI, Ligation of TEV protease with digested AraC induction system (NgoMIV and HindIII) and digested pJC354_ssTorA_XhoI_CS-TEV_NheI_BlaFL (HindIII and BamHI)
Preparing glycerol stocks of pUP089
Time: 2011-09-08
Investigators: Jessica
Materials:
- overnight cultures of pUP089
- glycerol
Method:
- 700 µl culture
- 300 µl glycerol
- stored in -80°C blue iGEM box
Output:
- pUP089 Aa, Jes, 08.09.11
- pUP089 Ab, Jes, 08.09.11
- pUP089 Ba, Jes, 08.09.11
- pUP089 Bb, Jes, 08.09.11
Ligation of mdnA-lib2 and mdnA-Age
Time: 2011-09-08
Investigators: Nicole
Purification of phages containing pPDV089
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- after over night incubation:
- centrifuge precipitaed phages: 5000 x g/ 45 min
- discard supernatant
- centrifuge again, 5000 x g/ 5 min
- remove supernatant carefully
- take pellet in 1 ml TBS
- move in 1.5 ml Eppi
- centrifuge again: 17000 x g/10 min
- supernatant in a new Eppi with 200 µl PEG-NaCl (mix!)
- incubate on ice for 60 min
- centrifuge precipitated phages: 17.000 x g/ 10 min (4°C)
- resuspend pellet in 300 µl TBS
- centrifuge:17 000 x g/ 10 min (4°C)
- supernatant in a fresh eppi= purified phages
Further tasks:
- measure concentration of phages
Glycerol stocks of phages containing pPDV089
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Method/Materials:
- 300 µl phages in TBS
- 750 µl 86 % glycerol
- store at -80°C
Further tasks:
- measure concentration of phages
Concentration of phages containing pPDV089 measured by nanodrop
Investigators: Sandrina
Aim: control if geneIII-mdnA will be expressed on the phage
Results:
- OD 269: 0,235
- OD 320: 0,010
- phages per ml:
Ph/ml = (Abs 269- Abs 320) * 6*10^16/ bp (phagemid)
- bp (phagemid) = 10945
Ph/ml= 1,23*10^12
Further tasks:
- measure concentration of phages by serial dilution
plasmid pereperation of pARWIII to control deletion of kanamycin gene, pSB1C3 containing mdnA, geneIII or mdnA/geneIII
Investigators: Sandrina, Sabine
Aim: control deletion of kanamycin gene in created pARWIII, control ligation of mdnA, geneIII and mdnA/geneIII into pSB1C3
Method/Materials:
plasmid preperation:
protocol 5.1 of the NucleoSpin Plasmid Kit
Further tasks:
test digestion
Antibody detection with blotted membranes
Investigators: Sandrina
Aim: control if geneIII-myc-mdnA is presented on the phages
Method/Materials:
- incubate blocked membrane for 1 h with primary antibody (anti -c-myc- antibody) in TBS with 5% milk powder
- wash 3x 10 min with TBS buffer
- incubate for 1 h with secondary antibody
- wash 3x 10 min with TBS buffer
- develop membranes with ECL- Kit
Results:
- signals were only seen at positive controls and one negative control
Further tasks:
90th Labday 2011-09-09
Preparation of samples for HPLC analysis
Time: 2011-09-09
Investigators: Jessica, Katharina
Material:
- pellets of pARW089 and pARW071 samples (from 2011-09-06)
- 100% methanol
- 5& methanol
- sterile water
- Sep-Pak Cartridges
- speed vac
Method:
- pellet was resuspended in 5 ml of sterile water
- the resuspended sample was sonicated for 5 min (3 sec on, 3 sec off)
- centrifugation for 10 min @ 11000 rpm
- supernatant was transfered to Sep-Pak Plus C18 Cartridges, that were equilibrated with 2 ml of 100% methanol
- cartridges were washed with 2 ml water
- samples were load
- cartridges were washed with 2 ml of 5% methanol
- samples were eluted with 2 ml of 100% methanol
- samples were put in a speedvac so that the methanol can evaporate
Counting of colonies on plates (pUP089+lib2)
Time: 2011-09-09,7:30
Investigators: Nadine, Nicole
Aim: count number of colonies on plates
Material:
- agar plate (kan): 10x pUP089+lib2, Nadja, 7.9.11, resistance: kan, from 2011-9-8, Nad/Nic
Results:
| plate | # of colonies | picked colonies |
| 1 | 61 | I clone E and F |
| 2 | 38 | II clone C and D |
| 3 | 87 | III clone A and B |
| 4 | 47 | IV clone G and H |
| 5 | 33 | IV clone I and J |
| 6 | 55 | |
| 7 | 37 | |
| 8 | 70 | |
| 9 | 28 | |
| 10 | 36 | |
| sum | 492 |
- example plates
Conclusions:
- 492 colonies
- some colonies will be picked for test sequencing (clone A-J)
- repeat fillin, ligation and transformation for more mutants
Glycerol stocks of pSB1C3+mdnDE K1, K2 and K5
Time: 2011-09-09, 8:00
Investigators: Nadine
Sequencing of pSB1C3+mdnD/ABC
Time: 2011-09-09, 8:00
Investigators: Nicole
Transformation of XL1 with different constructs
Time: 2011-09-09,
Investigators: Nadine, Nicole
Materials:
- XL1-Blue
- ligation products (2011-9-8):
- mdnA-lib2
- mdnA-Age
- mdnABCDE from pARW071
- antibiotics from stock:
- chloramphenicol
- kanamycin
- ampiciline
- LB-Agar
Method:
- ligation products:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 30 min on ice,
- heat shock 60 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (20 ml Agar and 20 µl antibiotic (see above) per plate)
- storage over night at 37°C (start: 15:00)
Further tasks:
- Picking clones for overnight culture (except for library 2)
- Producing glycerol stocks
- count colonies of pUP089_lib2 and collect them w/ LB
Output:
- Plates w/ XL1 blue:
- 10 x pUP089+mdnA-lib2 (on kan plates) + control
- 10 x pARWIII+mdnA-Age (on amp plates) + control
- pSB1C3+mdnABCDE from pARW071 (on cm plates) + control
Survival-Test of one PreScission clone( SG9-P10) and one Tev-clone (SG13-T6)
Investigators: Paul, Stefan, Sascha
Aim: Test whether TorA-detection device functions
- SG13-T6 contains two silent mutations and one single amino-acid exchange (N-->D)
- SG9-P10 contains different mutations:...
Plates:
30x, 15x for each clone
- 2x Cm (25 µg/ml)
- 2x Cm + 1mM IPTG
- 2x Cm + 1mM Ara
- 2x Cm + Amp (50 µg/ml)
- 2x Cm + Amp (200 µg/ml)
- 2x Cm + 1mM Ara + Amp (50 µg/ml)
- 2x Cm + 1mM Ara + Amp (100 µg/ml)
- 2x Cm + 1mM Ara + Amp (200 µg/ml)
- 2x Cm + 1mM Ara + Amp (400 µg/ml)
- 2x Cm + 1mM Ara + Amp (800 µg/ml)
- 2x Cm + 1mM Ara + 1mM IPTG + Amp (50 µg/ml)
- 2x Cm + 1mM Ara + 1mM IPTG + Amp (100 µg/ml)
- 2x Cm + 1mM Ara + 1mM IPTG + Amp (200 µg/ml)
- 2x Cm + 1mM Ara + 1mM IPTG + Amp (400 µg/ml)
- 2x Cm + 1mM Ara + 1mM IPTG + Amp (800 µg/ml)
Method:
- cells from glycerolstocks (-80°C) were inoculated in 5 ml LB-Medium (Cm = 25µg/ml). Then diluted and incubated until OD=0.002
- 100µl of cells with OD=0.002 were plated on prepared plates
Results:
Survival screening have to be repeated, because there is no way to compare numbers of clones on plates (dual induction of protease via AraC and ssTorA_CS_blaFL construct via IPTG) with number of clones on the right controll plates (singel induction with IPTG to check the export via TAT-pathway of beta lactamase.
No resistance against ampicillin without induction with IPTG. Dual-induction with arabinose and IPTG does not lead to a dramatical reduced number of clones.
Further Tasks:
- Repeat the survival test with singel induction via IPTG as comparison for the cell death based on reduced export of beta lactamase into the periplasm of E.coli
- repeat survival test at 37 °C for both protease to check influence of creation of inclusion body of TEV protease at 37°C and influence of export capacity of beta lactamase via TAT-pathway
Assembly PCR of mutated PreSciccon protease
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Stefan, Sebastian
Aim: Assembly of mutated PreSciccion
Materials:
- PreSciccion protease fragments P5 and P6 --> PreSciccion protease (total mutated)
- Primer 1: f_Pre_AraFusion_NgoMIV
- Primer 2: r_Pre_iGEM_BamHI
- Phusion polymerase kit by NEB
Methode:
PCR
- Template: 2 µL (1µl of each fragment after sizes were checked and fragments got cleaned up with PCR clean up kit from Macherey-Nagel (Nucleo Spin Kit))
- Nucleotides: 1 µL of 10 mM ready to use dNTP mix
- 10 µL 5 x Phusion HF Buffer
- 2,5 µL primers = 25 pmol absolute (2,5 µL of each primer, see list below)
- 31,5 µL of pure water
- 0,5 µL Phusion HF polymerase
Program:
- Denat: 30 sec 98°C
- 30x
Denat: 10 sec 98°C
Anneal/extend: 30 sec 72°C
- Final Extend: 10min 72°C
Further Tasks:
PCR purification, analytical Gel to check sizes of the amplified fragments. Digest and ligation.
Digestion of NgoMIV_TEV-Protease_iGEM_BamHI, NgoMIV_PreScission-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV fragments and pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5 and pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Paul, Sascha, Stefan
Materials:
NgoMIV_TEV-Protease_iGEM_BamHI: 66,2 ng/µl
NgoMIV_PreScission-Protease_iGEM_BamHI: 91,7ng/µl
HindIII_iGEM_AraC_NgoMIV: 48,3 ng/µl
pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (contains TEV cleavage site): 470 ng/µl
pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5 vector (contains PRE cleavage site): 270,2 ng/µl
Digestion protocol:
1: NgoMIV_TEV-Protease_iGEM_BamHI:
- 10µl NgoMIV_iGEM_TEV-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
2: NgoMIV_PreScission-Protease_iGEM_BamHI:
- 10µl NgoMIV_PreScission-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
3: HindIII_iGEM_AraC_NgoMIV:
- 10µl HindIII_iGEM_AraC_NgoMIV
- 1µl HindIII
- 1µl NgoMIV
- 5µl 10x buffer = NEB4
- 0.5µl 100x BSA
- 32.5µl pure water
- =50µl
4: pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5:
- 4µl pUP189-pJC354-NheI-TEV-Xho_blaFL_GGH5
- 1µl HindIII
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 39.5µl pure water
- =50µl
5: pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5:
- 8µl pUP189-pJC354-NheI-143C-Xho_blaFL_GGH5
- 1µl HindIII
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 37.5µl pure water
- =50µl
-->The reaction was allowed to proceed for 2h at 37°C!
Ligation of NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI or NgoMIV_iGEM_TEV-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV into pJC354-NheI-143C-Xho_blaFL_GGH5 or pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Sebastian, Sascha, Stefan
Aim:
- 1. Triple-ligation of NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI (573bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-143C-Xho_blaFL_GGH5 vector (~4700bp) to create pUP_SG2_TorA_CS-PreScission_bla_AraC-PreSciss
ion
- 2.Triple-ligation of NgoMIV_iGEM_TEV_iGEM_BamHI (760bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (~4700bp)to create pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
Calculation of volumes to be used with: [http://www.gibthon.org/ligate.html ligation calculator] with 1:1 molar ratio
Materials:
PreScission:
1:
- 1 µL NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI fragment
- 1 µL AraC fragment
- 6 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
- 1 controls: As 1 but with water instead of fragment
TEV:
- 1 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment
- 1 µL AraC fragment
- 6 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
- 1 control: As 4 but with water instead of fragment
TEV backbone with PreScission:
- 1 µL NgoMIV_iGEM_PreScission-Protease_iGEM_BamHI fragment
- 1 µL AraC fragment
- 6 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
PreScission backbone with TEV:
- 1 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment
- 1 µL AraC fragment
- 6 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
Used method:
ligation at room temperatur for 1h
Further task: Transformation of XL1blue cells with ligation products
Miniprep from overnight cultures T I/II, pre CS,PC, TEV-CS-1, P1
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Niels
Aim: isolate DNA & sequencing
Samples for miniprep
- T I
- T II
- pre-CS
- TEV-CS
- P1
- PC
Materials
miniprep by using Kit :
- NucleoSpin Plasmid
Nanodrop measurement
| Sample | concentration [ng/µl] | 260/280 | 260/230 |
| T I | 164,6 | 1,87 | 2,04 |
| T II | 182,7 | 1,88 | 2,07 |
| pre-cs | 263,1 | 1,86 | 2,13 |
| TEV-cs | 181,8 | 1,87 | 2,07 |
| PC | 150,5 | 1,85 | 2,05 |
| P1 | 357,6 | 1,86 | 2,19 |
futher task :
- sequencing
Sequencing of miniprep (09-09-2011) - T I/II, pre CS,PC, TEV-CS-1, P1
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Niels, Stefan
Samples: miniprep from 09-09-11
- T I
- T II
- TEV-cs
- pre-cs
- P1
- PC
- each:
- 1400 ng DNA
- ad 20 µl steril water
sequenced by GATC
Transformation of TEV and PreScission
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Sebastian, Sascha, Stefan
Aim:
Materials:
Heat shock protocol
Used method:
picking clones
Test digest of pSB1C3+mycN/C
Time: 2011-09-09
Investigators: Jessica
Aim: prove of Insert (mdnAmycN/C)
Materials:
- pSB1C3+mycN clones J1-J5, Niels, 9.9.11
- pSB1C3+mycC clones J1-J5, Niels, 9.9.11
- AvaII, PstI-HF (NEB)
- Buffer 4 (NEB, 10x)
- water
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnAmycN | 2596 | AvaII, Pst-HF | 4 | 225, 942, 1439 |
| mdnAmycC | 2597 | AvaII, Pst-HF | 4 | 225, 942, 1440 |
Digestion protocol
- 2 µl DNA
- 0.5 µl AvaII
- 0.5 µl PstI-HF
- 2 µl 10x buffer
- 15 µl pure water
- total: 20µl
- 37°C overnight
Agarose gel
| lane | Sample | Volume in µl |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
| 1 | mycN clone 1 | 20 + 4x loading dye |
| 2 | mycN clone 2 | 20 + 4x loading dye |
| 3 | mycN clone 3 | 20 + 4x loading dye |
| 4 | mycN clone 4 | 20 + 4x loading dye |
| 5 | mycN clone 5 | 20 + 4x loading dye |
| 6 | mycC clone 1 | 20 + 4x loading dye |
| 7 | mycC clone 2 | 20 + 4x loading dye |
| 8 | mycC clone 3 | 20 + 4x loading dye |
| 9 | mycC clone 4 | 20 + 4x loading dye |
| 10 | mycC clone 5 | 20 + 4x loading dye |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
Conclusions:
- digestion did not work out in a satisfying way
Further tasks:
- new restriction enzyme digestion
- sequencing
Miniprep of overnight cultures + pSB1C3+mycN/mycC
Time: 2011-09-09, 14:30
Investigators: Niels
Samples for miniprep
- pSB1C3+mycN J1
- pSB1C3+mycN J2
- pSB1C3+mycN J3
- pSB1C3+mycN J4
- pSB1C3+mycN J5
- pSB1C3+mycC J1
- pSB1C3+mycC J2
- pSB1C3+mycC J3
- pSB1C3+mycC J4
- pSB1C3+mycC J5
Materials
miniprep by using Kit :
- NucleoSpin Plasmid
Nanodrop measurement
| Sample | concentration [ng/µl] | 260/280 | 260/230 |
| mycN-1 | 189,7 | 1,87 | 2,46 |
| mycN-2 | 95,3 | 1,86 | 1,58 |
| mycN-3 | 132,0 | 1,85 | 2,57 |
| mycN-4 | 130,6 | 1,84 | 2,17 |
| mycN-5 | 152,8 | 1,86 | 2,46 |
| mycC-1 | 143,0 | 1,86 | 2,57 |
| mycC-2 | 92,5 | 1,88 | 2,11 |
| mycC-3 | 105,3 | 1,84 | 1,78 |
| mycC-4 | 109,5 | 1,84 | 1,98 |
| mycC-5 | 124,5 | 1,84 | 2,15 |
futher task :
- sequencing
Sequencing of pSB1C3+mdnDE/mycN/mycC
Time: 2011-09-09, 15:30
Investigators: Nicole, Niels, Jessica
Samples: miniprep from 09-09-11
- pSB1C3+mdnDE K1 (primers: VR2, sf_mdne_1_n (# 9), sf_mdne_2_1 (# 10), sf_mdne_3_1 (# 11))
- pSB1C3+mdnDE K2
- pSB1C3+mdnDE K5
- pSB1C3+mycN J1
- pSB1C3+mycN J2
- pSB1C3+mycN J3
- pSB1C3+mycN J4
- pSB1C3+mycN J5
- pSB1C3+mycC J1
- pSB1C3+mycC J2
- pSB1C3+mycC J3
- pSB1C3+mycC J4
- pSB1C3+mycC J5
- each:
- 1400 ng DNA
- ad 20 µl steril water
sequenced by GATC
Picking clones for overnight cultures of Lib 2 for confirmation
Investigators:Katharina
Time: 2011-09-09
Materials:
- agar plates with Lib2 clones
- LB medium
- kanamycin
Method:
- picking 10 clones
- Lib2 clone A-J
- incubate over night in 37°C shaker (200 rpm)
Output:
- 10 liquid cultures Lib2
Further tasks:
- miniprep
- sending for sequencing
Prepare liquid culture for mdnA foc2
Investigators:Katharina
Time: 2011-09-09
Materials:
- agar plates with Lib2 clones
- LB medium
- kanamycin
Method:
- using 5 ml of LB media (without antibiotic) for sluicing down the colonies from the first plate
- transferring the media to the next plate
- procedure for all 10 plates
- incubate over night in 37°C shaker (200 rpm)
Output:
- 1 liquid cultures mdnA foc2
Further tasks:
- miniprep
Picking clones for overnight cultures of pSB1C3+mdnABCDE with and without T7-promotor
Investigators:Katharina
Time: 2011-09-09
Materials:
- agar plates of pSB1C3+mndABCDE with T7 promotor and pSB1C3+mdnABCDE without T7-promotor
- LB medium
- chloramphenicol
Method:
- picking 4 clones for pSB1C3+mdnABCDE without T7-promotor
- picking 2 clones for pSB1C3+mdnABCDE with T7-promotor
- incubate over night @ 37°C (200 rpm)
Output:
- 4 liquid cultures of pSB1C3+mdnABCDE without T7-promotor
- 2 liquid cultures of pSB1C3+mndABCDE with T7
Further tasks:
- miniprep
- sending for sequencing
Preparing overnight cultures of expression backbone-clones
Investigators:Katharina
Time: 2011-09-09
Materials:
- agar plates of expression backbones
- pSB1A3_YFP_Ara
- pSB1A3_YFP_Lac
- pSB1K3_CFP_Ara
- pSB1K3_CFP_Lac
- pSB1K3_YFP_Lac
- LB medium
- ampicillin, kanamycin
Method:
- picking 1 clone for each construct into 10 ml LB media+antibiotic
- incubation overnight @ 37°C (200 rpm)
Output:
- 5 liquid cultures of different expression backbones
PCR: mdnABCDE
Time: 2011-09-09
Investigators: Nadja
Materials
- vector: pARW071 (1); pARW089 (2*)
- Long Enzyme Mix (+MgCl2) Buffer Fermentas
- Long Enzyme Mix Polymerase Fermentas
- dNTPs (NEB)
- water
- Primer:
| # | Primer |
|---|---|
| 84 | pf_mdnABCDE089_NotI_SpeI_30.08. |
| 83 | pf_mdnABCDE_T7_EcoRI_NotI |
| 82 | r_mdnABCDE_iGEM |
Protocol:
1. General composition
- 2 µl vector: pARW071 (1); pARW089 (2*) (diluted 1:100)
- 1 µl dNTPs (10 mM)
- 3 µl Primer forward (10 µM)
- 3 µl Primer backward (10 µM)
- 5 µl buffer
- 0.3 µl Ploymerase
- 35,7 µl water
- total volume: 50 µl
2. Composition
- pARW071 (1): 84, 82
- pARW089 (2*)-T7: 83, 82
- pARW089 (2*): 84. 82
3. PCR programs
- IGLONG2
Further task
- gel verification
- purification
test digestion to control if pSB1C3 is containing mdnA, geneIII or mdnA/geneIII
Investigators: Sandrina, Steffi
Aim:control ligation of mdnA, geneIII and mdnA/geneIII into pSB1C3
Method/Materials:
- 5 clones from pSB1C3 with mdnA (chloramphenicol), 5 clones from pSB1C3 with geneIII (chloramphenicol), 5 clones from pSB1C3 with mdnA/geneIII (chloramphenicol):
- 4 µl vector DNA (58- 120 ng/µl)
- 2 µl buffer 3
- 0,2 µl BSA
- 0.5 µl XbaI
- 0.5 µl PstI
12.8 µl water
incubate for 1 h at 37°C
Results:
Loading of gels
| lane | Sample | Volume in µl | Expected insert size in bp |
| M | marker, DNA ladder mix Fermentas | ||
| 1 | free | ||
| 2 | mdnA-geneIII fusion gene in pSB1C3, clone 1 | 12 | ca. 700 |
| 3 | mdnA-geneIII fusion gene in pSB1C3, clone 2 | 12 | ca. 700 |
| 4 | mdnA-geneIII fusion gene in pSB1C3, clone 3 | 12 | ca. 700 |
| 5 | mdnA-geneIII fusion gene in pSB1C3, clone 4 | 12 | ca. 700 |
| 6 | mdnA-geneIII fusion gene in pSB1C3, clone 5 | 12 | ca. 700 |
| 7 | geneIII in pSB1C3, clone 1 | 10 | ca. 500 |
| 8 | geneIII in pSB1C3, clone 2 | 10 | ca. 500 |
| 9 | geneIII in pSB1C3, clone 3 | 10 | ca. 500 |
| 10 | geneIII in pSB1C3, clone 4 | 10 | ca. 500 |
| 11 | geneIII in pSB1C3, clone 5 | 10 | ca. 500 |
| 12 | mdnA in pSB1C3, clone 1 | 12 | ca. 180 |
| 13 | mdnA in pSB1C3, clone 2 | 12 | ca. 180 |
| 14 | mdnA in pSB1C3, clone 3 | 12 | ca. 180 |
| 15 | mdnA in pSB1C3, clone 4 | 12 | ca. 180 |
| 16 | mdnA in pSB1C3, clone 5 | 12 | ca. 180 |
Further tasks:
sequencing of clones
Send clones 2-6 (geneIII-mdnA in pSB) for sequencing
Investigators: Sandrina, Steffi
Aim:control ligation of mdnA/geneIII into pSB1C3
Method/Materials:
- Primer: VR
- 20 µl with 70 ng/µl DNA
Further tasks:
check alignment
test digestion to control if kanamycin resistance gene is knocked out of pARWIII
Investigators: Sandrina, Steffi
Aim:control knock out of kanamycin on pARWIII
Method/Materials:
- 5 clones:
- 4 µl vector DNA (100- 300ng/µl)
- 2 µl buffer 3
- 0,2 µl BSA
- 0.5 µl NsiI
- 13.3 µl water
incubate for 1 h at 37°C
Results:
Loading of gels
| lane | Sample | Volume in µl | Expected insert size in bp |
| M | marker, DNA ladder mix Fermentas | ||
| 1 | pARWIII, clone 1 | 10 | no insert band |
| 2 | pARWIII, clone 2 | 10 | no insert |
| 3 | pARWIII, clone 3 | 10 | no insert |
| 4 | pARWIII, clone 4 | 10 | no insert |
| 5 | | 10 | no insert | - |
Further tasks:
sequencing of clones
91th Labday 2011-09-10
miniprep of Lib2 clones for confirmation
Time: 2011-09-10,9:00
Investigators: Niels, Nadine, Katharina
Materials:
- 10 overnight cultures (Lib2 clone A-J)
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| Lib2 clone A | 886.7 |
| Lib2 clone B | 979.7 |
| Lib2 clone C | 823.5 |
| Lib2 clone D | 815.5 |
| Lib2 clone E | 824.8 |
| Lib2 clone F | 987.7 |
| Lib2 clone G | 682.4 |
| Lib2 clone H | 683.4 |
| Lib2 clone I | 5.3 |
| Lib2 clone J | 767.5 |
Further Tasks:
- sending for sequencing
miniprep of mdnA foc2
Time: 2011-09-10,9:00
Investigators: Niels, Nadine, Katharina
Materials:
- 1 overnight culture
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnA foc2 | 610.0 |
Further Tasks:
- give it to the screening group
miniprep of pSB1C3+mdnABCDE with and without T7 promotor
Time: 2011-09-10
Investigators: Niels, Nadine, Katharina
Materials:
- 6 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnABCDE with T7-promotor clone 1 | 6.1 |
| mdnABCDE with T7-promotor clone 2 | 2.7 |
| mdnABCDE without T7-promotor clone 1 | 12.1 |
| mdnABCDE without T7-promotor clone 2 | 6.7 |
| mdnABCDE without T7-promotor clone 3 | 4.9 |
| mdnABCDE without T7-promotor clone 4 | 2.9 |
Further Tasks:
- doing new overnight culture and miniprep
Counting of colonies on plates (pUP089+lib2)
Time: 2011-09-09,10:00
Investigators: Nadine, Katharina, Niels
Aim: count number of colonies on plates
Material:
- agar plate (kan): 10x pUP089+lib2, Nic/Nad, 8.9.11, resistance: kan, from 2011-9-9, Nad/Kat
Results:
| plate | # of colonies |
| 1 | 78 |
| 2 | 92 |
| 3 | 65 |
| 4 | 55 |
| 5 | 67 |
| 6 | 80 |
| 7 | 116 |
| 8 | 87 |
| 9 | 66 |
| 10 | 35 |
| sum | 741 |
| control | 11 |
Conclusions:
- 741 colonies
- wash colonies from plates
- o.n. cultures and miniprep
- combine w/ preps from 2011-9-10 (Kat)
New test digest of pSB1C3+mycN/C
Time: 2011-09-10
Investigators: Niels, Nadine, Katharina
Aim: prove of Insert (mdnAmycN/C)
Materials:
- pSB1C3+mycN clones J1-J5, Niels, 9.9.11
- pSB1C3+mycC clones J1-J5, Niels, 9.9.11
- ScaI, XbaI (NEB)
- Buffer 3 (NEB, 10x)
- BSA
- water
Plan:
| insert | plasmid size in bp | enzymes | buffer | Expected size in bp |
| mdnAmycN | 2596 | ScaI, XbaI | 3 | 1024, 1570 |
| mdnAmycC | 2597 | ScaI, XbaI | 3 | 1024, 1570 |
Digestion protocol
- 1 µl DNA
- 0.5 µl ScaI
- 0.5 µl XbaI
- 2 µl 10x buffer
- 0.3 µl BSA
- 15.7 µl pure water
- total: 20µl
- 37°C for 1h
Agarose gel
| lane | Sample | Volume in µl |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
| 1 | mycN clone 1 | 20 + 4x loading dye |
| 2 | mycN clone 2 | 20 + 4x loading dye |
| 3 | mycN clone 3 | 20 + 4x loading dye |
| 4 | mycN clone 4 | 20 + 4x loading dye |
| 5 | mycN clone 5 | 20 + 4x loading dye |
| 6 | mycC clone 1 | 20 + 4x loading dye |
| 7 | mycC clone 2 | 20 + 4x loading dye |
| 8 | mycC clone 3 | 20 + 4x loading dye |
| 9 | mycC clone 4 | 20 + 4x loading dye |
| 10 | mycC clone 5 | 20 + 4x loading dye |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
Conclusions:
- digestion did not work out in a satisfying way
Test Expression Backbones: pSB1K3_Ara/Lac_CFP
Time: 2011-09-10,11:00
Investigators: Nadine, Katharina, Niels
Aim:
prove of promotors (Lac/Ara)
Materials/Methods
- start cell grow in fresh media
| time | time of grow [min] | OD600 Ara_CFP | OD600 Ara_CFP control | OD600 Lac_CFP | OD600 Lac_CFP control | |
| 11:00 | 0 | 0,147 | 0,147 | 0,121 | 0,130 | |
| 11:30 | 30 | 0,156 | 0,160 | 0,132 | 0,139 | |
| 12:00 | 60 | 0,182 | 0,183 | 0,149 | 0,152 | |
| 12:30 | 90 | 0,212 | 0,203 | 0,172 | 0,174 | |
| 13:00 | 120 | 0,260 | 0,257 | 0,210 | 0,207 | |
| 13:30 | 150 | 0,323 | 0,331 | 0,259 | 0,260 | |
| 14:00 | 180 | 0,406 | 0,416 | 0,326 | 0,370 | |
| 14:30 | 210 | ind. | ind. | 0,370 | 0,403 | |
| 15:00 | 150 | ind. | ind. | 0,446 | 0,466 |
- after inducing by IPTG/ARA - samples where measured for 2h every 15 min
- result negativ
further task
- repeat experiment
Glycerol stocks: Expression Backbones
Time: 2011-09-10,11:00
Investigators: Nadine, Katharina, Niels
Material:
- 300 µl DNA of the following expression backbones
- pSB1A3_YFP_Ara
- pSB1A3_YFP_Lac
- pSB1K3_CFP_Ara
- pSB1K3_CFP_Lac
- pSB1K3_YFP_Lac
- mdnA Lib-foc2
- 700 µl 86 % glycerol
- store at -80°C
Oligo-Fillin for mdnA-library 2 (repetition) and for Phage-display-mdnA-library (carrying AgeI restriction site)
Investigators: Niels, Nadine, Katharina
Time: 2011-09-10
Aim: Production of libraries carrying mutated mdnA, one for phage-display (carrying ageI restriction site)
Material:
- oligos (25 mM, HPLC purified, ordered by Sigma Aldrich):
- o_mdnA_library, 76
- o_focused_library_AgeI, 85
- 10x Klenow buffer
- dNTPs (10 mM)
- H2O
Method:
- Reaction mix (total volume: 50 µl)
- 3 µl fw-oligonucleotide (o_mdnA-library, 76)
- 3 µl rev-oligonucleotide (o_focused_library_AgeI, 85)
- 5 µl dNTPs
- 5 µl Klenow buffer
- 34 µl H2O
- Reaction conditions
- Program: ORIGAMI1
- after finishing program ORIGAMI1:
- addition of 1 µl Klenow-fragment
- incubation 1 hour, 37°C
Results:
- hybridized oligos for production of mdnA-library
Output:
- AgeI-library, named AgeI
Further tasks:
- PCR purification
- restriction enzyme digestion using SfoI and AgeI, also of pARWIII
Restriction enzyme digestion of ParWIII and fill-in-reaction of Libary 2
Investigators: Katharina, Niels, Nadine, Steffi
Time: 2011-09-10, 20:30
Material:
digest of Fill-in-Reaction of Libary 2 (2011-09-10) :
- 22,5 µl DNA (fill-in-product
- 2,5 µl Age I
- 2,0 µl Sfo I
- 3µl Buffer 3
- 37°C over night
further task :
- purification by Kit
digestion of ParWIII:
- 6,5 µl template (pARWIII)
- 3 µl Buffer 4
- 2.5 µl Age I
- 2.0 µl Sfo I
- 16 µl water
- 37°C over night
further task :
- purification by gel
further tasks:
- ligation
- transformation
Restriction enzyme digestion of pSB1C3 for ligation with mycN/C
Investigators: Nadine, Katharina, Niels
Time: 2011-09-10, 20:00
Material:
purified PCR products (2011-09-03) :
- mycC: 4.2 ng/µl
- mycN: 15.2 ng/µl
digestion of pSB1C3:
- 6 µl DNA (235.0 ng/µl (2011-06-22))
- 3 µl Buffer 4
- 1.5 µl XbaI
- 2.0 µl PstI
- 0.5 µl BSA
- 11.5 µl water
- 37°C over night
further tasks:
- purification
- ligation
- transformation
Restriction enzyme digestion of pSB1C3 for control
Investigators: Niels, Nadine, Katharina, Steffi
Time: 2011-09-10, 20:30
Material:
pSB1C3(2011-06-22) :
- concentration: 235,0 ng/µl
digestion of pSB1C3 with Bgl I:
- 2 µl DNA (235.0 ng/µl (2011-06-22))
- 1 µl Buffer 3
- 0.5 µl BglI
- 6.5 µl water
- total volume 10 µl
- 37°C over night
- expected fragments[bp]:
- 1169
- 962
- 314
- 199
digestion of pSB1C3 with SacI, TaqI:
- 2 µl DNA (235.0 ng/µl (2011-06-22))
- 1 µl Buffer 4
- 0.5 µl SacI
- 0,5 µl TaqI
- 0,3 µl BSA
- 5,7 µl water
- total volume 10 µl
- 37°C over night
- expected fragments [bg]:
- 1446
- 648
- 548
| lane | Sample | Volume in µl |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
| 1 | pSB1C3 (BglI) | 10 + 2 loading dye |
| 2 | pSB1C3 (SacI und TaqI) | 10 + 2 loading dye |
Picking clones for overnight cultures of pSB1C3+mdnABCDE
Investigators:Katharina, Steffi, Niels, Nadine
Time: 2011-09-10 20:45
Materials:
- agar plates of pSB1C3+mndABCDE (2011-09-09)
- LB medium
- chloramphenicol
Method:
- picking 5 clones for pSB1C3+mdnABCDE (2011-09-09)
- incubate over night @ 37°C (200 rpm)
Output:
- 5 liquid cultures of pSB1C3+mdnABCDE without T7-promotor
Further tasks:
- miniprep
- sending for sequencing
Preparing overnight cultures of expression backbone-clones
Investigators:Katharina, Niels, Nadine, Steffi
Time: 2011-09-10 19:30
Materials/Method:
- liquid culture :
- pSB1K3_CFP_Ara
- pSB1K3_CFP_Lac
- 10 ml LB medium
- 10 µl kanamycin
Output:
- 2 liquid cultures of expression CFP backbones with different promotors
Transformation of mdnABCDE + pSB1C3
Time: 2011-09-10 21:30
Investigators:Niels, Nadine, Katharina, Steffi
Aim: Transformation of E.coli cells with mdnABCDE + pSB1C3 (with and without T7) - ligation
Materials:
- competent E. coli cells (XL1-Blue, 2011-08-29)
- ligation products: mdnABCDE + pSB1C3
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 20 min on ice,
- heat shock 90 sec at 42°C,
- incubation 10 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking(750 rpm) for 60 min,
- centrifuge 30 min at 2000 x g
- discard the supernatant (till ~100µl media)
- plating on LB plate with appropriate antibiotic (Cm)
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Output:
to be continued....
Colony PCR of plated TEV and PreScission containing colonies
Investigators: Stefan, Paul, Sebastian
Aim:
get dna for BioBricks
Used Primer:
Tev:
f_TEV_iGEM, r_TEV_iGEM_BamHI
PreScission:
f_Pre_iGEM, r_Pre_iGem:BamHI
Method:
- 2,5µl of each primer (10µM) = 5µl
- 5µl 10y polymerase buffer
- 2µl 25mM MgCl2
- 1µl dNTP mix
- 0,5µl Taq-Polymerase (GenAxxon)
- 36,5 µl H2O
=50µl
Colonies were picked with a 20µl tip, dipped into PCR batch, and then plunged into 1ml LB to grew cells from picked colony (from these 1ml batches overnight cultures will be inoculated in case of positive clones).
- 10 colonies were picked for TEV and 10 colonies for PreScission protease (see entry from 01.09.2011) = 20 PCR batches.
PCR Program:
Initial denat = 3min 94°C
25x
denat: 2min 10sec 94°C
anneal: 2min 10sec 70°C
extend: 1min 72°C
final extend: 10min 72°C
- The PCR products were resolved on a 1% analytical agarose gel
Results:
no bands!
Further Tasks:
repeat it!
Digest of TEV, PreSciccion, AraC and different Vectors (pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL, pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL and pSB1C3)
Investigator: Nadine, Sebastian
Material:
Enzymes: (all pruchased from NEB)
- HindIII
- BamHI
- NgoMIV
- XbaI
- PstI
DNA-Fragments:
- mutated TEV protease
- mutated PreSciccion protease
- AraC
- pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL
- pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL
- pSB1C3
NEBuffer 4, 3 and 1
BSA from NEB
Method:
All digestion batches got a total volume of 10µl and were incubated for 2 h at 37°C:
- 1µl Buffer
- 1µl each Enzyme
- 0,1 µl BSA if required
- 7 or 6,9 µl DNA
Both protease (TEV and PreSciccion) got digested with BamHI+NgoMIV or NgoMIV+ PstI, AraC was digested with NgoMIV+HindIII or NgoMIV+XbaI. Both vector which containing the cleavage site for the Protease got digested with HindIII+BamHI, the vector pSB1C3 was digested with XbaI and PstI.
Output:
Digested fragements:
- NgoMIV_TEV_BamHI
- NgoMIV_TEV_PstI
- NgoMIV_PreSciccion_BamHI
- NgoMIV_PreSciccion_PstI
- HindIII_AraC_NgoMIV
- XbaI_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
- BamHI_ pJS354_ssTorA_XhoI_CS-Pre_NheI_blaFL_HindIII
- PstI_pSB1C3_XbaI
Further Tasks:
Clean up of all fragments via agarose gel and extraction of digested fragment from the agarose gel, ligation of fragments and teransformation into E.coli XL1 blue.
Gelextraction of digested fragements, ligation and transfomration of ligation products into E.coli XL1 blue
Investigator:
Sebastian, Sascha, Stefan
Aim:
Getting digested fragments purified, to get lost of small DNA fragments, we don't need.
Materials:
Digested fragements:
- NgoMIV_TEV_BamHI
- NgoMIV_TEV_PstI
- NgoMIV_PreSciccion_BamHI
- NgoMIV_PreSciccion_PstI
- HindIII_AraC_NgoMIV
- XbaI_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
- BamHI_ pJS354_ssTorA_XhoI_CS-Pre_NheI_blaFL_HindIII
- PstI_pSB1C3_XbaI
Methode:
Seperate digestion batches over an 1% agarose gel, cut of the bands with the right size and perform a gel purification with Nucleo spin II kit from Macherey-Nagel.
Output.
Digested and purified fragments of:
- NgoMIV_TEV_BamHI
- NgoMIV_TEV_PstI
- NgoMIV_PreSciccion_BamHI
- NgoMIV_PreSciccion_PstI
- HindIII_AraC_NgoMIV
- XbaI_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
- BamHI_ pJS354_ssTorA_XhoI_CS-Pre_NheI_blaFL_HindIII
- PstI_pSB1C3_XbaI
Further tasks:
Ligation of fragments into digested vector and transformation of E.coli XL1 blue with ligation products. Picking clones, performing a colony PCR, overnight cultures from all positive clones and creation of an glycerol stock culture and miniprep of those positive plasmids for sequencing.
Ligation of digested fragments of TEV protease, PreSciccion protease and different vectors
Investigators: Stefan, Sascha, Paul
Materials:
Fragments:
- NgoMIV_TEV_BamHI
- NgoMIV_TEV_PstI
- NgoMIV_PreSciccion_BamHI
- NgoMIV_PreSciccion_PstI
- HindIII_AraC_NgoMIV
- XbaI_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
- BamHI_ pJS354_ssTorA_XhoI_CS-Pre_NheI_blaFL_HindIII
- PstI_pSB1C3_XbaI
QuickLigase purchased by Fermentas
QuickLigase 10x Buffer purchased by Fermentas
Methodes:
Standard ligation protocol from Fermantas for QuickLigase Kit. Ratio between vector and both fragments (protease and AraC-induction system) where 3:1:1.
Reaction batches:
Every batch contains 1µl QuickLigase 10x Buffer, and 0,5 µl QuickLigase (both by Fermentas).
- Batch 1:
- NgoMIV_TEV_BamHI
- HindIII_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-TEV_NheI_blaFL_HindIII
- Batch 2:
- NgoMIV_PreSciccion_BamHI
- HindIII_AraC_NgoMIV
- BamHI_ pJS354_ssTorA_XhoI_CS-Pre_NheI_blaFL_HindIII
- Batch 3:
- NgoMIV_TEV_PstI
- XbaI_AraC_NgoMIV
- PstI_pSB1C3_XbaI
- Batch 4:
- NgoMIV_PreSciccion_PstI
- XbaI_AraC_NgoMIV
- PstI_pSB1C3_XbaI
Every batch was incubated for 10 min at 37°C. After 10 min, all batches where put on ice.
Output:
- 4 different ligation batches
Further tasks:
- Transformation of E.coli XL1 blue with ligation products, colony PCR to get to know, which clones are positive and for all positive clones: mini-prep of plasmid DNA, sequencing and preparation of glycerol stock clutures.
Transfromation of E.coli XL1 blue with ligation products
Investigators: Sascha, Paul, Sebastian
Aim:
transformation of E.coli XL1 blue with ligation batcheshttp://www.syntbio.net/iGEM/wiki2011/index.php/Labjournal_september#Ligation_of_digested_fragments_of_TEV_protease.2C_PreSciccion_protease_and_different_vectors
Materials:
- 4 different ligation batches http://www.syntbio.net/iGEM/wiki2011/index.php/Labjournal_september#Ligation_of_digested_fragments_of_TEV_protease.2C_PreSciccion_protease_and_different_vectors
- competent E.coli XL1 blue cells
Method:
Standard transformation protocol using heat shock.
Output:
4 different E.coli cultures transformed with different ligation batches.
Further tasks:
plating of the transformed E.coli cultures on LB agar plates containing chloramphenicol.
Plating of transformed E.coli XL1 blue
Investigators: Stefan, Paul, Sebastian
Aim:
getting single positive E.coli XL1 blue colonies transformed with the different ligation batches http://www.syntbio.net/iGEM/wiki2011/index.php/Labjournal_september#Ligation_of_digested_fragments_of_TEV_protease.2C_PreSciccion_protease_and_different_vectors
Method:
100 µl of the transformed cultures where plated out on lB agar plates containing chloramphenicol, the rest of the transformed cultures where centrifuge at 4000 g for 3 min. The supernatant was discarded and the pellet re suspended in 100 µl LB media, which was also plated out on a new LB agar plate containing chloramphenicol. All agar plates where incubated over night at 37°C.
Further tasks:
picking clones for colony PCR and mini prep of plasmid DNA, also establishing of glycerol stock cultures and preparing plasmids for sequencing.
 "
"