Team:NTNU Trondheim/Journal Modeling
From 2011.igem.org
m (→Wednesday 27/7) |
m (→Thursday 11/8) |
||
| Line 209: | Line 209: | ||
== Thursday 11/8 == | == Thursday 11/8 == | ||
| - | Tried to | + | Tried to learn python, and make a script that plots the output from the tau - leap program. It doesn't work at all, who says python is easy?!? |
| - | + | ||
== Friday 12/8 == | == Friday 12/8 == | ||
Revision as of 10:01, 19 September 2011

Journal for Modeling Group
|
|
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
You can find an overview of the modeling here.
Wednesday 22/6
Introductory lectures in systems biology was held by Marius Eidsaa. Different approaches were discussed and it was decided to work with two different models: Bayesian Networks and systems of Ordinary Differential Equations, and possibly extend it to Stochastic Differential Equations.
Friday 24/6
Dizzy was installed and we started to look at how to simulate Michealis Menten reactions.
Monday 27/6
We continued where we ended last week, with having lectures and to get aquinted with different software.
Thursday 30/6
A program which solves ordinary differential equations numerically using the Runge Kutta method of order 4 and 5 was made in Matlab. Using expressions based on proteins inhibiting mRNA and having the level of ppGpp fixed the reponse of the system could be found.
Tried to find suitable parameters, but found this to be hard. Contacted the wetlab group and got help to find the processes which cause inhibation. This can be used for modeling in Dizzy.
The Bayesian model started to take form, and is ready to be implemented in C++.
Friday 1/7
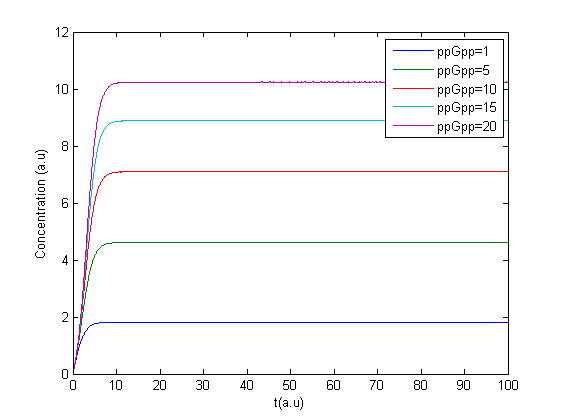
The Matlab program was tested to see if it gave reasonable results. Minor errors were corrected and the first results were obtained, due to a lack of accurate parameters the units on the axis are arbitrary. Here is the Matlab code:
tspan=[0;20]; % defines the start and stop
x=10.0; %the initial conentration of ppGpp
%the initial concentrations of LacI mRNA, LacI, mCherry mRNA and mCherry
y0=[20;20;0;0];
[T Y]=ode45(@F,tspan,y0);
plot(T,Y(:,2),T,Y(:,4),'r')
ppGpp=num2str(x);
title(['ppGpp=',ppGpp])
legend('LacI','mCherry','Location','East')
xlabel('t(a.u)')
ylabel('Concentration (a.u)')
Where F is a function defined by:
function Y = F(t,y)
Y=zeros(4,1);
x=10.0;
k1=[5e-4;5e-4];
k_prime=[20.0;20.0];
d1=[1;1];
n=[1;1];
k2=[20;20];
d2=[20;20];
Y(1)=k1(1)+k_prime(1)/(1+x^n(1))-d1(1)*y(1);
Y(2)=k2(1)*y(1)-d2(1)*y(2);
Y(3)=k1(2)+k_prime(2)/(1+y(2)^n(2))-d1(2)*y(3);
Y(4)=k2(2)*y(3)-d2(2)*y(4);
end
The resulting plot is shown in this figure.
Monday 4/7
The program was modified to make it possible to have several values of ppGpp in one run making it possible to see the response to different values of ppGpp, see figure.
The theory for the Bayesian model seems to be in place, see figure , some of the implementation has already been done in C++.
Tuesday 5/7
Started to implement the model in Dizzy and looking at the relationship between this model and the model implemented in Matlab. The model in Dizzy involves many more processes, if these are assumed to be fast the species involved can be assumed to be in equilibrium and be replaced with a steady state value. The current model in Dizzy involves 10 processes, while the Matlab model only have four.
Thursday 7/7
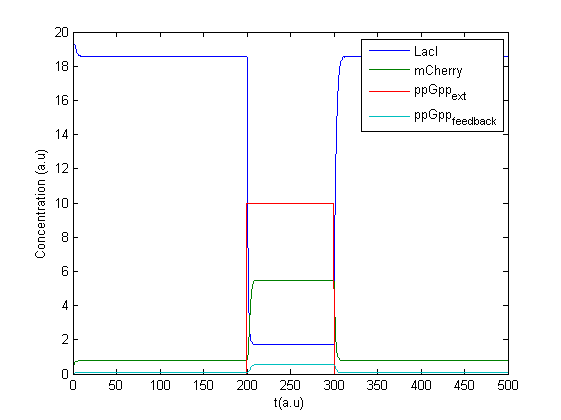
A new program was made in Matlab to treat the concentration of ppGpp as an input signal. A possible feedback effect was also included as production of mCherry might lead to increase in ppGpp. The concentration of ppGpp was split into two parts, one external controlled and one caused by feedback , see figure.
Friday 8/7
Derived expressions for the steady state concentrations and compared it to the results obtained in Matlab.
We were given an introductory lecture in molecular biology and biotechnology by Gunvor, to get an understanding of the work beeing done at the lab.
Monday 11/7
After a team meeting we was given a Master Thesis by Sigurd Johansen describing a toogle switch. We started to expand the ODE model to include more reactions than we have done so far. Both to be aware of possible limitations of the current model and to get a clearer understanding of the fundamental processes involved.
Started to investigate how to model and solve stochastic differential equations, and how to derive them from physical properties.
Wednesday 13/7
The Master Thesis includes a lot more step then currently included in our model and from these step a set of four equations are obtained. Processes currently not considered in our model includes: Several ways of inhibation (i.e both one or two ppGpp kolecules might inactivate the rrnB promoter) dimerization of the Lac repressor, several steps during transcription.
Thursday 14/7
We contacted the lab group to clarify the way ppGpp inhibits the rrnB promotor and was informed that it affects the RNAp and not the promotor in itself and can therefore also affect pLac.
The theory behind stochastic differential equation was investigated further.
Wednesday 20/7
We contacted Autodesk to explore the possibility of using Maya and the mMaya toolkit to visiualize the system. We also visited the lab to see how gelelectrophoresis was performed.
Thursday 21/7
We recieved mail from Autodesk, installed mMaya and explored the possibility of creating animations.
Friday 22/7
A simple animation of a green bacteria (or something resembling that) turning red was made in Maya.
Wednesday 27/7
Meeting with prof. Almaas.
We decided to drop the bayesian model (see here), and focus on stochastic models.
Thursday 28/7
Dizzi was installed on one of the calculations server at NTNU, Dept. of Mathematics.
An electronic mail was send to see if another iGEM team needed help (requirement for gold medal).
Friday 29/7
Looked into regression models for stress response, using non-linear regression with heteroskedasticity (from mass spectrometry??). Think that Kernel smoothing estimation might be the best approach.
Saturday 30/7
Kernel smoothing might be a bad idea, found out how to do smoothing B - splines with unequal weighting in R, which is just what we need. The weight should be inverse proporsjonal with the variance at each point.
Monday 1/8
Sozi was installed in the computer lab, and we looked into the presentation and poster. How to make that one great? Using Inkscape.. Looked at some of the poster/presentation from last year, and found out what NOT to do, but not what to do.
Searched for reasonable parameters for the dizzy simulation.
Tuesday 2/8
Found out how to use loops in Dizzy, which makes it easy to vary parameters and start condition. Experimented with storing data from dizzy and made a script in matlab which plots the data.
Wednesday 3/8
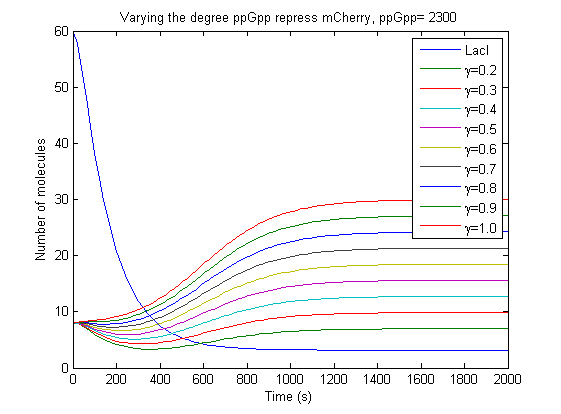
Began to take advantage of the features developed yesterday, started to study what effect a possible inhibation of mCherry by ppGpp could have, see figure.
Made a short presentation in sozi, to show how it should be done in Amsterdam
Thursday 4/8
Met with prof. Eivind Almaas, to discuss further progress, started to extend the tau-leap program with complex prameters.
Friday 5/8
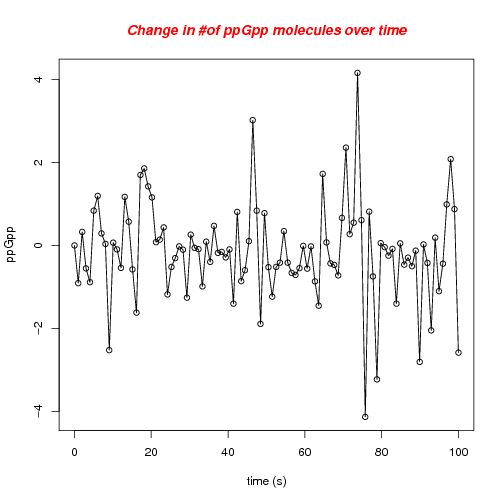
Found a reasonable way of simulating ppGpp in the cell as a "restricted" random walk, using a mixture of a uniform and a exponential random variable.
Looked into how to plot the output from the C++ program on tau leap, decide that python might be best (but never used it before)
Saturday 6/8
Writing on the model page... and some basic python trail; "Hello World!"
Monday 8/8
Throw the old poster overboard, and started on a new one.
Tuesday 9/8
Used Dizzi on the calculation server, produced weird results, no idea why.
Wednesday 10/8
Continued to work on the new poster
Thursday 11/8
Tried to learn python, and make a script that plots the output from the tau - leap program. It doesn't work at all, who says python is easy?!?
Friday 12/8
Parameter testing..
 "
"