Team:METU-BIN Ankara/Project
From 2011.igem.org
| Line 47: | Line 47: | ||
<div id="act_content"> | <div id="act_content"> | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | ul {list-style-type: | + | ul {list-style-type:disc;list-style-image:none;} |
</style> | </style> | ||
<h2>Project</h2> | <h2>Project</h2> | ||
Revision as of 19:05, 16 September 2011
Project
Introduction
Synthetic biology is a rising new field. Every year igem getting more important for newbies at the synthetic biology field. So each year number of the attending teams are increasing. As team number increases, submission of new biobricks are increasing too. All competitor are knowing that Partsregistry is a huge library for synthetic biologists. Although it contains precious informations for biologists, the parts are not organized very well. Therefore, while constructing devices, synthetic biologists have following difficulties in pre-experimental stage:
- searching the appropriate parts,
- deciding which part will work effectively with biobrick,
- finding the most accurate combination between the parts,
- constructing the most effectively working devices etc.
All these problems are time consuming and takes too much effort. Therefore this year main goal of METU-BIN 2011 project is to provide a web based tool that helps synthetic biologists at the pre-experimental step to design their genetic constructs using the biobricks according to their input and output parameters.
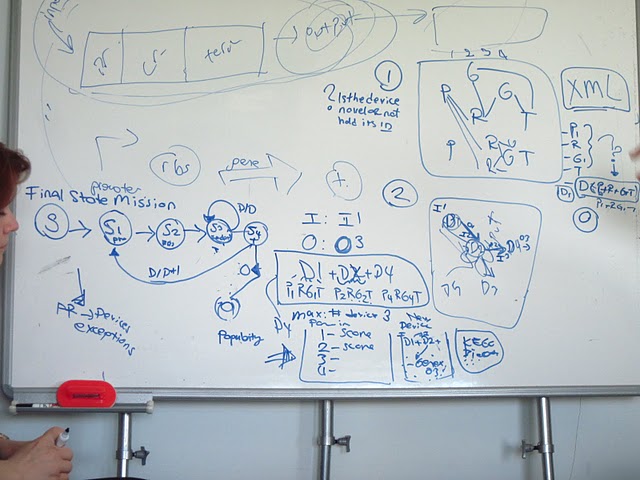
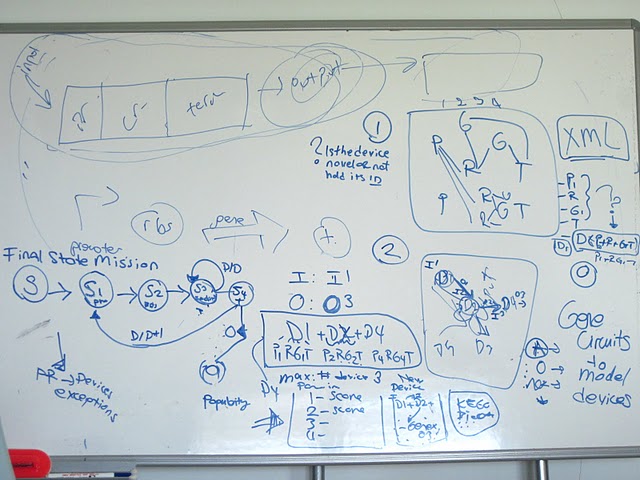
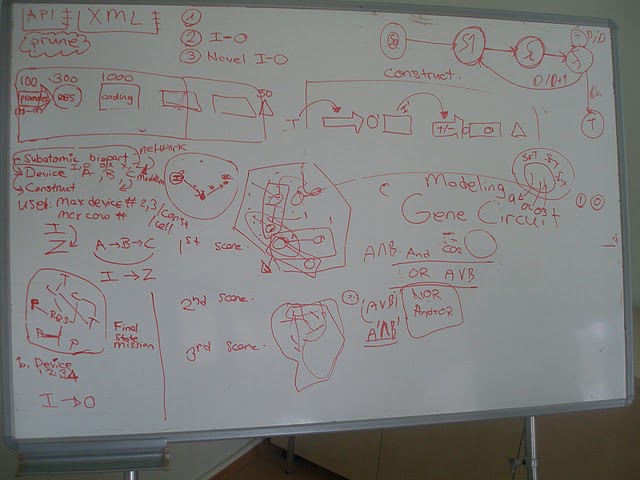
To help synthetic biologist at the problems they are facing we developed a program called M4B: Mining for Biobricks. Our software proposes a method which can be utilized in pre-experimental step as a supporter data mining tool. According to the user defined input and output, this software presents and scores all possible composite devices which are activated by the input and produce the requested output.
Aim
Main goal of our 2011 project is to provide a web based tool that helps synthetic biologists at the pre-experimental step to design their genetic constructs according to their input and output parameters.
- A network of all bioparts in 2011 distribution will be generated, which describes the functional relations between the subatomic bioparts.
- A search algorithm will be developed to reveal all possible device combinations for the user defined input and output within bioparts of 2011 distribution.
- Visualization tools will be applied for graphical representation of the results.
- A web-based user interface will be provided for the developed software.
 "
"