Team:BU Wellesley Software/G-nomeSurferPro
From 2011.igem.org
(Difference between revisions)
| Line 66: | Line 66: | ||
| - | <h5>Future Work:</h5> | + | <h5><a name=surferFuture></a>Future Work:</h5> |
In the future, G-nome Surfer will be expanded to include integration with the Clotho database and other tools like <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a>. Plans are also in place to expand annotations to include GenBank notes and publications. Information about the workspace will also be savable, with a session being preserved and re-accessible and selected publications lists being accessible outside the application through email or as .txt files. | In the future, G-nome Surfer will be expanded to include integration with the Clotho database and other tools like <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a>. Plans are also in place to expand annotations to include GenBank notes and publications. Information about the workspace will also be savable, with a session being preserved and re-accessible and selected publications lists being accessible outside the application through email or as .txt files. | ||
Revision as of 16:49, 12 September 2011
Computational Team
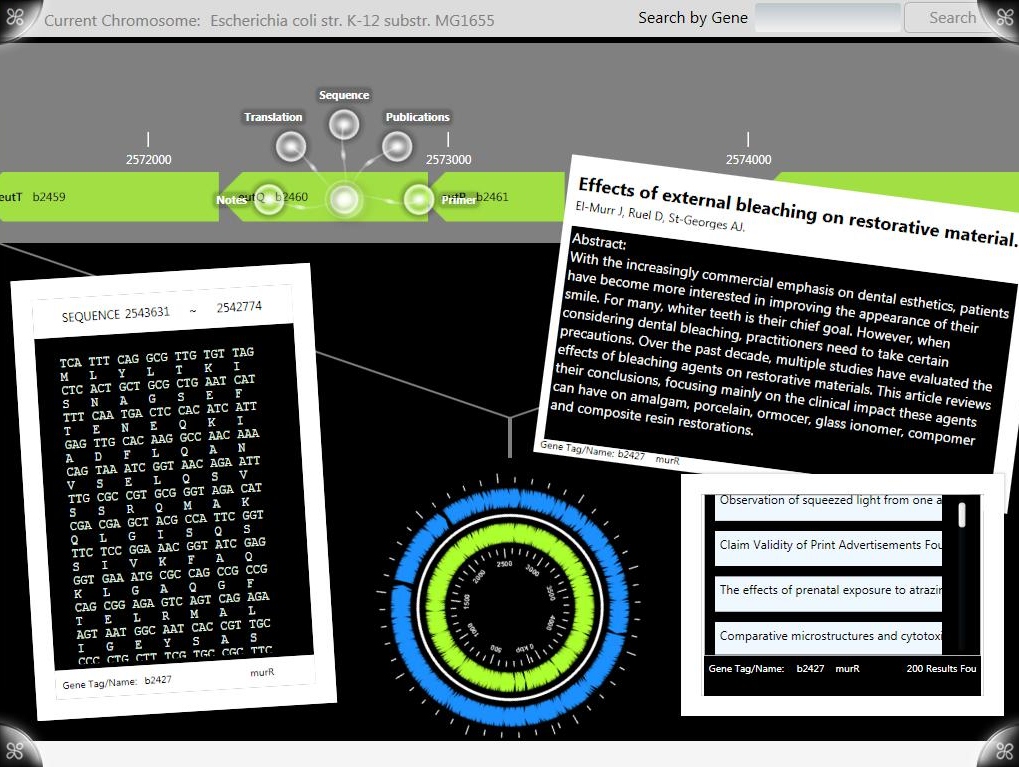
G-nome Surfer Pro is written in C# for the Microsoft Surface using the Surface SDK. State is managed using the MVVM Architecture and time consuming processes, like retrieving publications and running BLAST, are run in threads behind the main UI thread.
After selecting a genome from the tree menu the chromosome bar is populated with genes from the GenBank file. Another genome can be selected at anytime from the pull down search menu.
The chromosome bar is infinitely scrolling and position on the chromosome bar is tied into the orientation of the chromosome wheel through the MVVM architecture allowing manipulation of one to affect change of the other. GenBank files are parsed for sequence, translation and any notes, all of which are available as information objects, which are accessible from any gene on the bar. Users can take genes on the bar directly into the Optimus Primer surface application for primer design.
Information from any gene’s GenBank file are viewable as “ScatterViewItem” objects which can be moved and re-sized on the main “ScatterView” workspace. Users can view and annotate sequence/translation information by dragging and overlaying one over the other. All information objects can be moved to and from the extended desktop by dragging or tossing or erased by tossing them off the sides of the workspace. By moving objects between ScatterViews, users can view and organize data in a distraction free environment which can be pulled up over the main workspace or hidden from view.
Publications are populated in a scrollable list and can be dragged and dropped from the listBox to the main workspace or extended desktop through manipulation of the objects template. The PubMed search results for the gene are fetched and parsed for abstracts, authors, and publication data.
Future Work:
In the future, G-nome Surfer will be expanded to include integration with the Clotho database and other tools like Trumpet. Plans are also in place to expand annotations to include GenBank notes and publications. Information about the workspace will also be savable, with a session being preserved and re-accessible and selected publications lists being accessible outside the application through email or as .txt files.Ethical User Study practices:
Overview: G-nome Surfer Pro
Hypothesis Forming
G-nome Surfer Pro is an integrated environment for viewing prokaryotic genomic data and literature. Users can find genes on the chromosome wheel or search for a gene name or Genbank number, access Genbank notes and publications from Pubmed and view alignment and translation from the chromosome bar. The extended desktop provides distraction free workspace for information processing and hypothesis formation. The Microsoft Surface allows users to collaborate in pairs and teams on design and research tasks.

Demo Video
Wetlab
Lorem ipsum dolor sit amet, consectetur adipiscing elit. In et dictum leo. Maecenas porttitor augue nec arcu lacinia ultricies. Maecenas at dictum augue. Proin eget odio ac mi tristique scelerisque. Integer at
nisl nec purus laoreet condimentum sed semper dui. Duis feugiat, ligula
eu vehicula vulputate, sem nisl ornare neque, eu dictum lacus sapien
eu neque.
Integer at
nisl nec purus laoreet condimentum sed semper dui. Duis feugiat, ligula
eu vehicula vulputate, sem nisl ornare neque, eu dictum lacus sapien
eu neque.
Vestibulum gravida, turpis tempus suscipit euismod, ipsum mi tristique nibh, sed sagittis ante felis at urna. Suspendisse eu neque vitae lorem elementum elementum sit amet ac ligula. Mauris vestibulum Donec ac sapien erat. Proin id enim sed dolor suscipit laoreet vitae sit amet ligula. Nam ultricies orci vitae mauris egestas tincidunt. Vestibulum sit amet est dolor.
Results:
Ut eu nunc eget ante egestas egestas at eu metus. Integer quam justo, vehicula non sodales id, consequat vitae eros.Aenean egestas, ipsum sed fringilla porta, erat mi facilisis lectus, quis elementum quam felis nec elit.
 "
"
