Team:ETH Zurich/Biology/Detector
From 2011.igem.org
| Line 6: | Line 6: | ||
{|class="biologyTitle linkMap" border="0" cellspacing="0" cellpadding="3" align="right" style="border-radius:10px; text-align:center; white-space:nowrap; color: black;" | {|class="biologyTitle linkMap" border="0" cellspacing="0" cellpadding="3" align="right" style="border-radius:10px; text-align:center; white-space:nowrap; color: black;" | ||
|- | |- | ||
| - | |style="color:white; width: 3em; border-left: none;"|[[#Acetaldehyde Sensor|Acetaldehyde Sensor]] | + | |style="color:white; width: 3em; border-left: none;"|[[#Acetaldehyde Sensor:|Acetaldehyde Sensor]] |
| - | |[[#Xylene Sensor|Xylene Sensor]] | + | |[[#Xylene Sensor:|Xylene Sensor]] |
|} | |} | ||
|- | |- | ||
Revision as of 10:07, 11 September 2011
| Sensors |
| ||
| Overview about possible smoke sensors
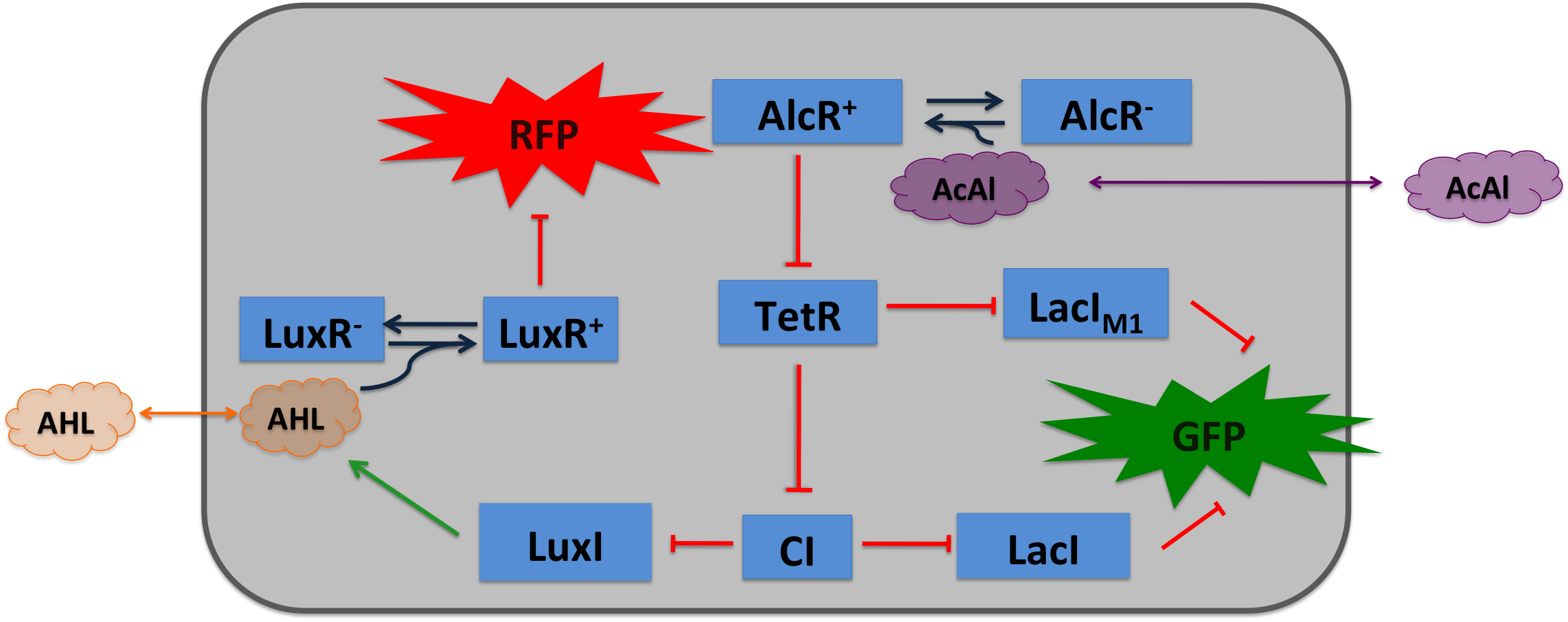
Acetaldehyde Sensor:
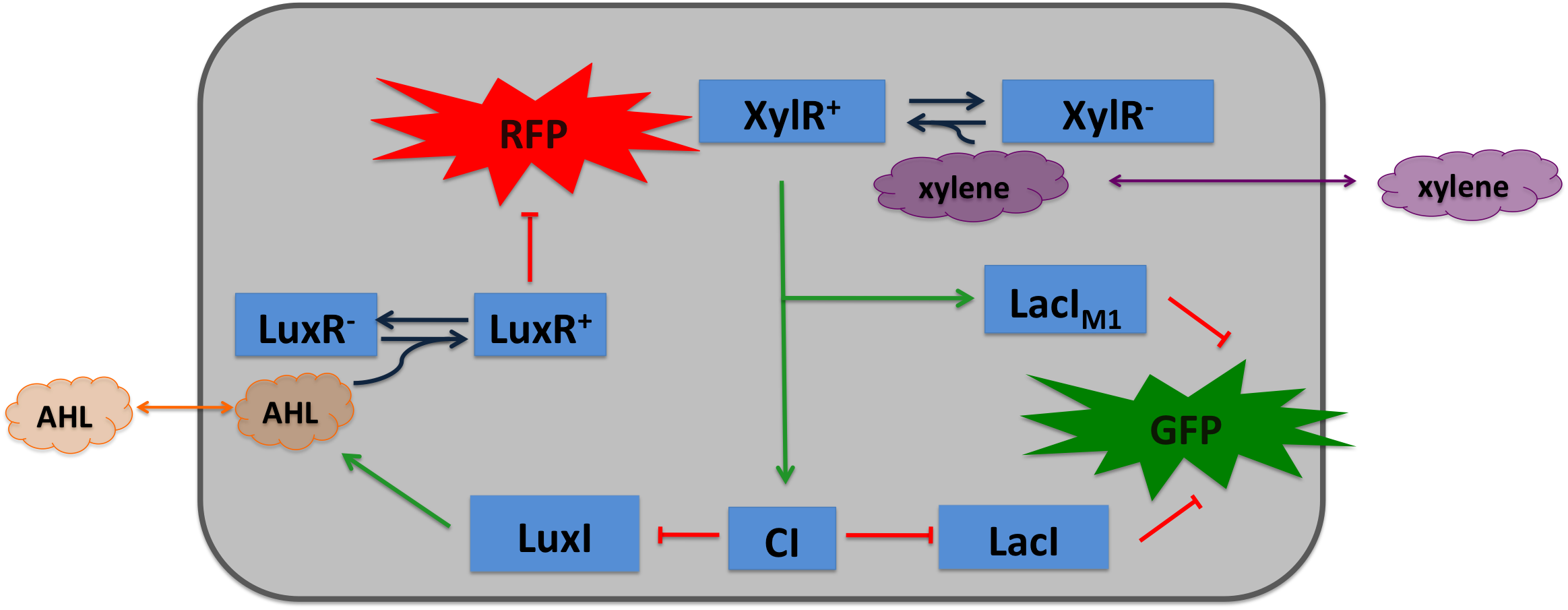
Xylene Sensor:
References[1] [http://www.ncbi.nlm.nih.gov/pubmed/2834622 R. Locklngton, C. Scazzocchio, D. Sequeval, M. Mathieu, B. Felenbok: Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans, Mol Microbiol., 1987, 1:275-81] [2] [http://aem.asm.org/cgi/content/abstract/64/2/748 Sven Panke, Juan M. Sánchez-Romero, and Víctor de Lorenzo: Engineering of Quasi-Natural Pseudomonas putida Strains for Toluene Metabolism through an ortho-Cleavage Degradation Pathway, Appl Environ Microbiol, 1998, 64: 748-751] [3] [http://aem.asm.org/cgi/content/abstract/64/6/2032 Sven Panke, Bernard Witholt, Andreas Schmid, and Marcel G. Wubbolts: Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120, Appl Environ Microbiol, 1998, 64: 2032-2043] | |||
 "
"