August 8
Testing for biofilms grown on 96-well plate and Erlenmeyer flasks with crystal violet
PCR of 16s
Transformation of I732019, I732005, J23119 from source plate in XL-10 gold.
Re-assembly of DDT gene using alternate method
GCMS of p450-degraded BPA samples
Obtain experimental strain (biofilm-forming) MG-1655 E. coli and grow overnight in SOC
Results
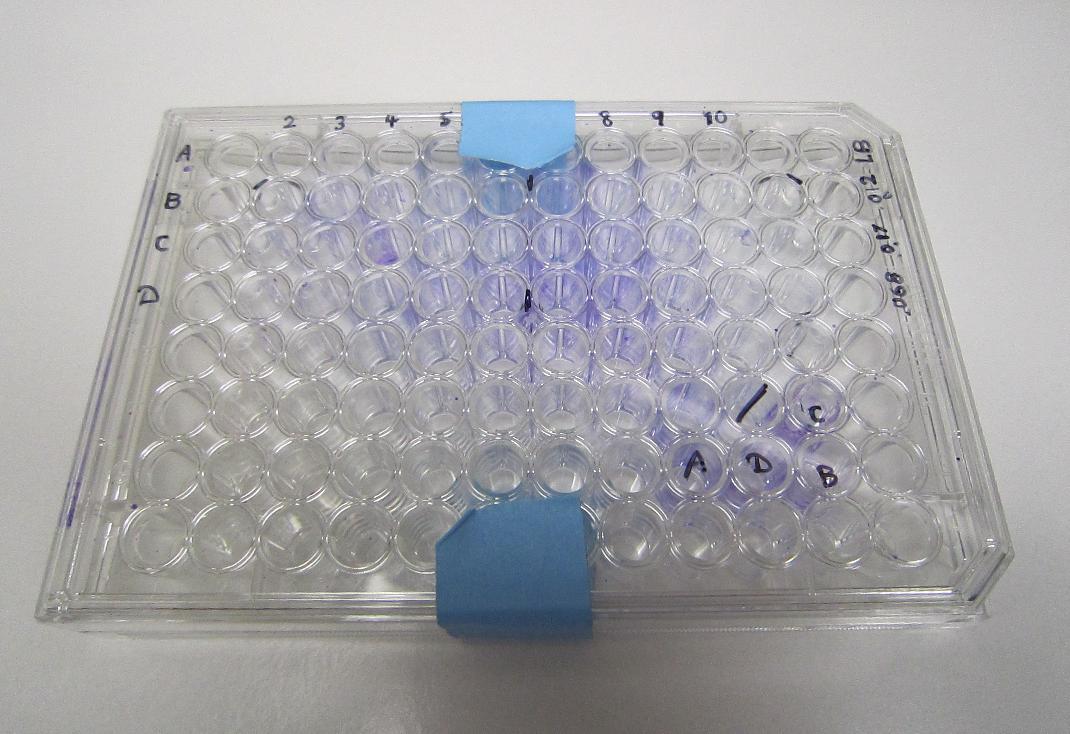
Bacteria-incubated well remained cystal violet stained after several rinses, indicating the presence of a biofilm, in contrast to the clear control well.
96-well plate with XL-10 biofilm grown for two days, visualized with 1% crystal violet
|
Glass beads with XL-10 biofilm grown for two days, visualized with 1% crystal violet
|
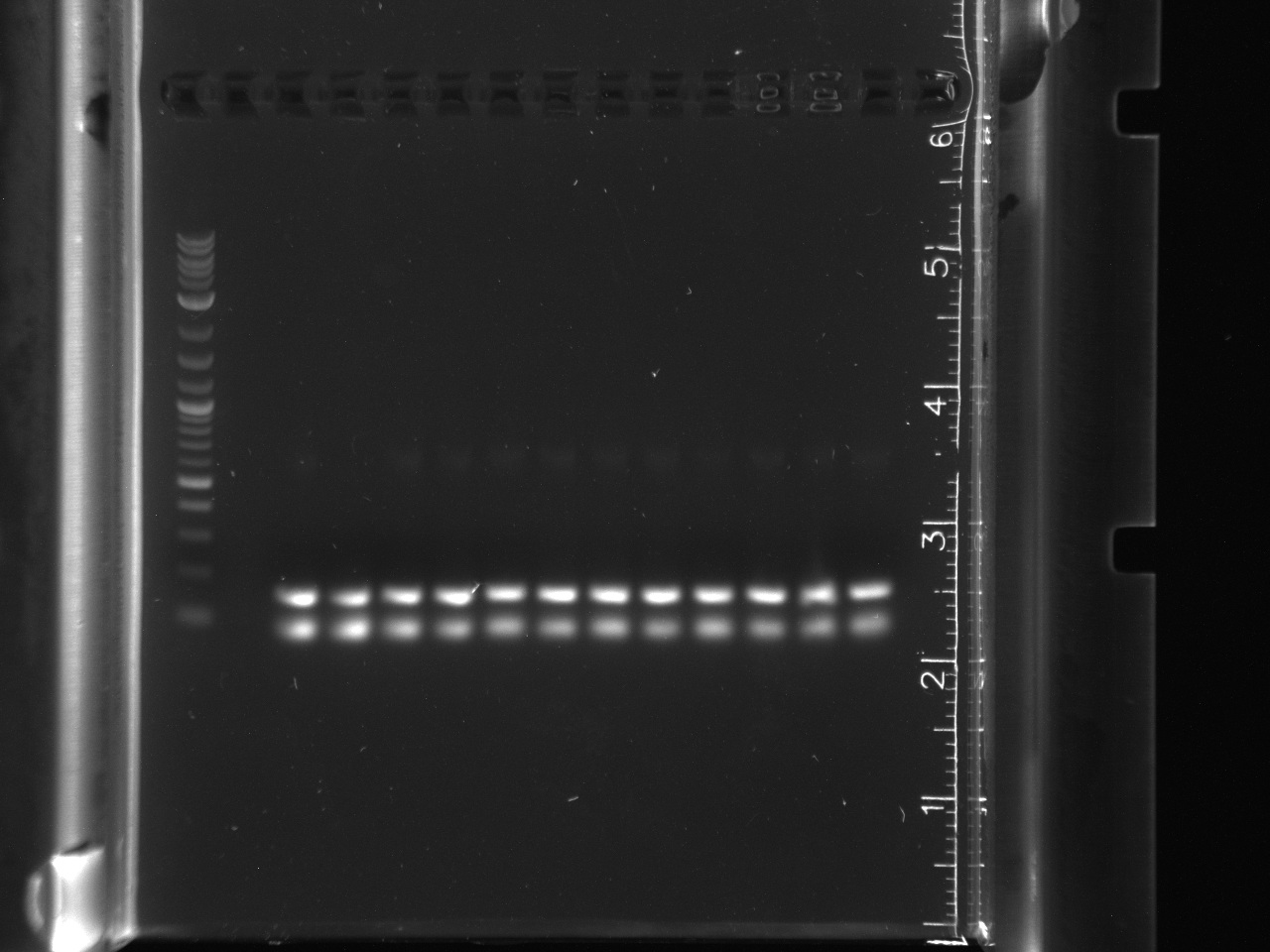
Got successful amplification of 16s in LA River sample 9-9. In both gels, the controls had no bands and all other sample lanes had no bands.
BPA sample was too diluted for GCMS; must concentrate first.
DDT assembly failed.
August 9
Evaporate-centrifuge BPA sample to concentrate for GCMS
Create glycerol stocks of experimental E. coli strain MG-1655 for biofilm experiments
Did 16s sequencing experiments with a Master Mix.
Gibson assembly of pNT004 with PCRd insert
CPCR of more pNT002 colonies with VF2 and VR primers.
Retransform I732005 and I732019 with top ten cells. Plated on kan plates and amp plates since plasmid has resistance to both.
Re-assembly of DDT gene using PIPE cloning and Gibson.
Results
I732019 and I732005 plates had no colonies.
Sequencing shows pNT003 and pNT002 colonies were self-ligated. But then what was the band on the cpcr gel?
DDT assembly failed.
16s gel had primer lengths, gene did not amplify.
August 10
Minimal media transfers
Started enrichment cultures of water samples from Baxter and BBB ponds.
Redid Gibson of pNT002, pNT003, pNT004 with pcrd inserts
Transformed cells with I732017 and K323089 for some source of lacZ
Re-assembly of DDT gene using PIPE cloning and Gibson, transformed into different cells.
Begin time-dependent biofilm optimization experiment with crystal violet stain.
Results
I732019 and I732005 plates had no colonies
pNT004 Gibson plates had no colonies
CPCR of pNT002 colonies had no insert bands
DDT assembly transformations showed colonies
August 11
Colony PCR of DDT gene colonies
Transformation of I732005, I732017, I732019 in new XL-10 cells
PCR of pSB4A5, pSB3K3, pNT002 insert, pNT003 insert. Tried PCA to assemble inserts more specifically.
Continue time-dependent biofilm optimization experiment with crystal violet stain.
Results
| Plate |
number of colonies |
| pNT002 + top ten |
3 |
| pNT002 - top ten |
1 |
| pNT003+ top ten |
1 |
| pNT003 - top ten |
0 |
| pNT004 + top ten |
1 |
| pNT004 - top ten |
1 |
| I372017 |
0 |
| K323089 |
0 |
| Name |
concentration (ng/ul) |
| pSB4A5 |
57.1 |
| pSB3k3 1 |
60.4 |
| pSB3k3 2 |
31.1 |
| pNT002 insert |
136.9 |
| pNT003 insert |
72.0 |
| pNT002 insert PCA |
15.7 |
August 12
Continue time-dependent biofilm optimization experiment with crystal violet stain.
PCR of 16s.
Results
Biobrick transformation plates
| Plate |
Colonies |
| I732005 |
0 |
| I732017 |
~200 |
| I732019 |
65 |

16s amplification, lane 1 NEB 2-log ladder
August 13
Results
| Plate |
Colonies |
| pNT002 + |
~320 |
| pNT002 - |
~800 |
| pNT003+ |
~500 |
| pNT003- |
~400 |
Crystal violet biofilm results:
| Starting OD |
8/10 |
8/11 |
8/12 |
8/13 |
| 5pm |
11pm |
11am |
5pm |
11pm |
11am |
5pm |
11pm |
11am |
5pm |
| 0.118 |
.065 |
.171 |
.114 |
.135 |
.266 |
.149 |
.133 |
.100 |
.408 |
.740 |
| 0.170 |
.065 |
.132 |
.082 |
.190 |
.159 |
.439 |
1.875 |
.106 |
.323 |
.545 |
| 0.068 |
.065 |
.106 |
.184 |
.198 |
.210 |
.121 |
.201 |
.145 |
.039 |
.881 |
|
Biofilm experiment 96-well plate
|
Biofilm experiment cuvettes
|
 "
"