Team:USC
From 2011.igem.org
(Difference between revisions)
| Line 66: | Line 66: | ||
<h1 style="font-family:Verdana;font-weight:700;">Project Description</h1> | <h1 style="font-family:Verdana;font-weight:700;">Project Description</h1> | ||
</tr> | </tr> | ||
| + | |||
| + | <tr> | ||
| + | <td> | ||
| + | <p> | ||
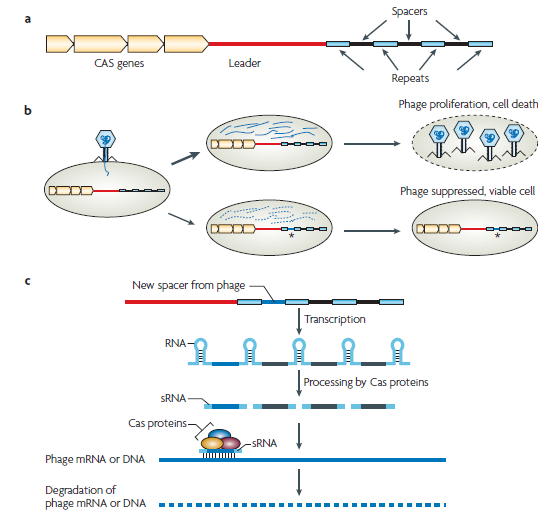
| + | Bacteria protect their genome and remove foreign DNA elements through the use of the clustered, regularly interspaced short palindromic repeat (CRIPSR) system. Recently discovered, this defense mechanism can protect microorganisms against bacteriaphages. The CRISPR system comprises a transcribed locus of repeats and spacers that is activated and made functional via a family of CAS proteins. The CRISPR repeats are short DNA sequences that are almost identical in size and sequence. Between a pair of repeats is a spacer, which if complementary to a DNA element of the invading bacteriaphage, facilitates the destruction of that sequence from the bacterial cell. As such, the CRISPR/CAS system is a primitive biological defense mechanism, similar in many ways to the process of adaptive immunity in eukaryotic immune systems. Plasmids, which are commonly used in molecular biology are also the source of horizontal gene transfer in the wild and facilitate the spread of antibiotic resistant genes among microorganisms. These plasmids can contain unique DNA sequences that could be targeted by the CRISPR/CAS system. We intend to exploit the CRISPR/CAS system to engineer a mechanism of plasmid curing and deactivation of antibiotic resistance genes in E. coli. | ||
| + | </p> | ||
| + | </td> | ||
| + | </tr> | ||
| + | |||
<tr> | <tr> | ||
<td style=" text-align:center; width:724px;"> | <td style=" text-align:center; width:724px;"> | ||
| Line 71: | Line 80: | ||
</td> | </td> | ||
</tr> | </tr> | ||
| + | |||
| + | |||
| + | |||
| + | |||
</table> | </table> | ||
</td> | </td> | ||
</tr> | </tr> | ||
</table> | </table> | ||
 "
"