Team:Harvard/Template:NotebookData4
From 2011.igem.org
(→Generated Final Chip Sequences) |
|||
| Line 276: | Line 276: | ||
**During this time, we created a function that will re-translate the sequences that the generator output. It compares the original helix with the re-translated helix to make sure that our reverse-translate works properly. | **During this time, we created a function that will re-translate the sequences that the generator output. It compares the original helix with the re-translated helix to make sure that our reverse-translate works properly. | ||
***This step went smoothly, and we verified that the sequences were reverse-translated properly. | ***This step went smoothly, and we verified that the sequences were reverse-translated properly. | ||
| - | **To make sure that the distributions generated were as expected, we made | + | **To make sure that the distributions generated were as expected, we made WebLogos of the helices generated(see below). |
*The output file (in the Dropbox: iGem > chip > final chip.csv) originally had the following headers: 'Target', 'Backbone #', 'Helix Sequence', 'Backbone Sequence', 'Nucleotide Sequence of Zinc Finger' | *The output file (in the Dropbox: iGem > chip > final chip.csv) originally had the following headers: 'Target', 'Backbone #', 'Helix Sequence', 'Backbone Sequence', 'Nucleotide Sequence of Zinc Finger' | ||
**We wanted to convert this information into FASTA format. | **We wanted to convert this information into FASTA format. | ||
Revision as of 15:53, 3 August 2011
Contents |
June 24
- Designed primer for testing HisB deletion, reuse His_Internal_R to test the band
Updated Closest Zif268 Fingers
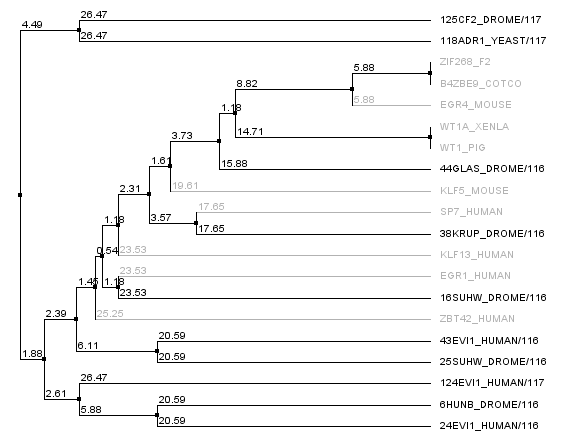
We realized that some of our "close non-zif268 fingers" were actually not all that close to Zif268, and so we went into the 88,000 zinc finger database and pulled out zinc fingers surrounding zif268. In fact, there were many, many, many zinc fingers that had identical sequences to the Zif268 F2 finger, and so we looked at sequences around it. The tree below shows the new non-zif268 backbones that are actually close to zif268 compared to our old set. The new set is in gray, the old set is in black. This gives us a potential seven more backbones to work with.
June 24th - Bioinformatics
Sequence Generation
We made some small updates to the sequence generator, based on the frequencies we noticed in the outputs of the tests we ran.
- We decided to only include pseudocounts for position 6 for 'CNN' and 'ANN.' Originally, 'CNN' and 'ANN' were using pseudocounts for all seven positions. However, this introduced a noticeable increase in amino acids, such as tyrosine (Y), that have been shown to occur rarely in zinc fingers (according to our data from OPEN and Persikov). Additionally, because tryosines occured so rarely in the data (11 times total in the open data set), we decided not to give tyrosine a pseudocount.
- We added the capability to prevent repeat backbone-helix combinations on the chip. That is, we wanted to make sure that the same exact zinc finger was not generated for different triplet inputs.
To test the sequence generator, we made two sets of 2000 sequences for GAA, then infographic-d the results. Comparing these with the images for OPEN and OPEN+Persikov shows that our generation follows the major themes of those datasets, but also introduces variation. The two generated sets also vary slightly from each other, which shows the influence of randomness on the generation.
| Disease | Target DNA Finger 1 | Helices in Zif268 Backbone | Helices in Zif268 Closely-Related Backbones | Helices in Zif268 Distantly-Related Backbones |
| Colorblindness (Bottom) | TGG | 5150 | 3000 | 1000 |
| Colorblindness (Top) | ATG | 3050 | 3050 | 3050 |
| Familial Hypercholesterolemia (Bottom) | GAC | 5150 | 3000 | 1000 |
| Familial Hypercholesterolemia (Top) | CTG | 3050 | 3050 | 3050 |
| Myc (Top198) | CTC | 3050 | 3050 | 3050 |
| Myc (Top981) | AAA | 3050 | 3050 | 3050 |
Table of target sequences and helix distribution across backbones
- Distribution: Zif268 : Zif268 similar : Zif 268 dissimilar
- Conservative distribution 56.3 : 32.8 : 10.9
- Riskier distribution 33.3 : 33.3 : 33.3
June 24th
pZE21G:
- reinoculated culture with 100µL of saturated solution, grew to mid-log, and made glycerol stock
- backbone PCR: ran E gel but no bands--PCR unsuccessful. We may need to use a different backbone for the zinc fingers.
Omega and Omega+Zif268:
- these were the only two PCR reactions from 6/22/11 to work
- PCR purified using Qiagen kit:
- omega: 6.1ng/µL, 260/280=1.83
- omega+Zif268: 11.3 ng/µL, 260/280=1.67
Lambda red recombination of selection system:
- reinoculated selection strain+pKD46 with 100µL of saturated solution
- just before mid-log (about 4 hours after inoculation) divided culture in half (1.5mL) and added either 37.5µL or 3.75µL of 20% arabinose solution (to try two different induction levels). Cultures grew for another hour.
- The rest of the procedure was the same as the 6/22/11 attempt but without the 42C water bath.
June 24th - Bioinformatics
Playing with Pseudocounts
Using CTC because of position 6's reliance on the CNN frequencies, we see what difference values of pseudocounts (if in the frequency table, the frequency of an amino acid is 0, bump it up to the psuedocount: ex. A = 0 becomes A = .015 with a psuedocount of .015) make. Pseudocounts are necessary for data that has small sample size - we could be missing out on working helices because a letter's frequency is 0 when it shouldn't be.
Various pseudocount (psu = ) values. Look at the 7th column, which is position 6 in the helix:
The variation from E being the top letter to A being top back to E is from a slight adjustment in how we add on psuedocounts: the 'new' way is a more proportional approach.
Notice how psu = 0 gives only the four letters found in our dataset, while psu > 0 adds in other letters, each with a small probability ranging from .5% to 2%.
The question is how much psu to add: less means we weight our (possibly flawed) data of proven zinc fingers more. Higher psu adds more randomness (variation) to our sequences, but some fraction of those sequences will not work.
List of Remaining Goals:
- Sort fingers by target
- Pick and assign primer sets
- Reverse translate fingers avoiding type II restriction enzymes and primers
- Append type II restriction enzyme and primer sequences to each finger
- Yay
June 25th-26th - Bioinformatics
This is the set of final target sequences with assigned forward and reverse primers (tags for PCR):
| Disease | Target Sequence | Forward Primer (5'-3' NOT REVERSE COMPLEMENT) | Reverse Primer (5'-3' NOT REVERSE COMPLEMENT) |
| Colorblindness | GCT GGC TGG | ATATAGATGCCGTCCTAGCG | AAGTATCTTTCCTGTGCCCA |
| Colorblindness | GCG GTA ATG | CCCTTTAATCAGATGCGTCG | TGGTAGTAATAAGGGCGACC |
| Familial Hypercholesterolemia | GGC TGA GAC | TTGGTCATGTGCTTTTCGTT | AGGGGTATCGGATACTCAGA |
| Familial Hypercholesterolemia | GGA GTC CTG | GGGTGGGTAAATGGTAATGC | ATCGATTCCCCGGATATAGC |
| Myc-gene Cancer | GGC TGA CTC | TCCGACGGGGAGTATATACT | TACTAACTGCTTCAGGCCAA |
| Myc-gene Cancer | GGC TGG AAA | CATGTTTAGGAACGCTACCG | AATAATCTCCGTTCCCTCCC |
June 27
Wet lab
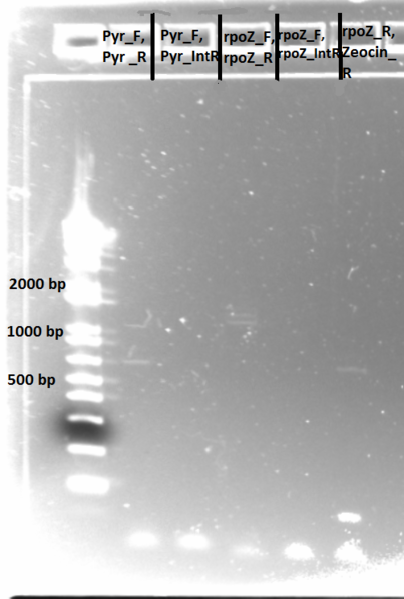
Sequencing PyrF, rpoZ loci:
- We will sequence these genes in the selection strain just to make sure they are knocked out, especially since it appears HisB is not.
- Picked a colony off ∆HisB∆PyrF∆rpoZ plate (6/21) and grew in 150µL LB plus tet in a 96 well plate for about 2 hrs at 37˚C
- diluted 1 in 20 and used 1µL as template in PCR with KAPA mastermix (see protocols for reagent amounts and parameters)
- annealing temp 65˚C, elongation time 1:15
- PyrF_F, PyrF_R primers
- PyrF_F, PyrF_internalR
- rpoZ_F, rpoZ_R
- rpoZ_F, rpoZ_internalR
- rpoZ_R, zeocin_R
- Run on E Gel to check PCR worked: bands are at the same sizes as the original genotyping gel.
- Tomorrow we will send samples to Genewiz for sequencing
Lambda Red recombination:
- The plates made from the recombination (6/24) did have colonies, but they were very small and took a long time to grow, and so they may not actually have the kan-ZFB insert. We will have to PCR the locus to see.
- Chose 8 colonies from each plate and grew at 30˚C in 150µL LB plus kan in a 96 well plate
- When our primers arrive, we will PCR the locus to check for the insert.
Selection system media:
June 27th - Bioinformatics
To Do for Today
- 100 sequences (and control), 2 each with the same F3 and F2, but different F1, from our test sequences [zif268, OZ123, OZ052, CoDA]✓
- Type II nuclease cut site sequences- put the binding sites into our oligos ✓
- Final backbones with helices ✓
- Programming stuff- Check to make sure there are no cut sites or primers in any of our backbone/helices combinations; check translation order (translates F1→F3)✓
100 Control Sequences
- See our Positive Control Sequences, updated June 28th
- Selected known binding zinc fingers from the CODA table that bind sequences similar to our target sequences
- All control helices from CODA were inserted into Zif268 F2 backbones and have been assigned a seventh primer tag separate from the tags given to the 6 target sequences.
Updated Target Sequences
One of our sequences from before was bad because the F3/F2 combo did not appear in the CODA table... faulty checking, my bad :(
Here is the newest table of target sequences:
| Disease | Target Range | Binding Site Location | Bottom Finger | Top Finger | Bottom AA (F3 to F1) | Top AA (F3 to F1) |
| Colorblindness | chrX:153,403,001-153,407,000 | 3666 | GTG GGA TGG | GAA GGG ACC | RNTALQH.QSAHLKR.####### | QDGNLGR.RREHLVR.####### |
| Familial Hypercholesterolemia | chr19:11,175,000-11,195,000 | 14001 | GGC TGA GAC | GGA GTC CTG | ESGHLKR.QREHLTT.####### | QTTHLSR.DHSSLKR.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 198 | GGT GCA GGG | GGC TGA CTC | VDHHLRR.QSTTLKR.RRAHLQN | ESGHLKR.QREHLTT.####### |
| Myc-gene Cancer | chr8:128,938,529-128,941,440 | 981 | GGA GAG GGT | GGC TGG AAA | QANHLSR.RQDNLGR.TRQKLET | EKSHLTR.RREHLTI.####### |
- Green cells are our target sequences.
Cut Site Design
- See our Cut Site Design page
- We left in one proline (P) between the linker and the starting FCQ... of finger 2, but as this proline is the last AA of the OPEN linker (TGEKP) and occurs before the beta sheet in every zinc finger in Zif268 (see zif268's sequence on its [http://www.pdb.org/pdb/explore/remediatedSequence.do?structureId=1AAY PDB page])
- This configuration also allows the library to be used at any finger position because proline ends the OPEN linker.
Updates on the program
The program appears to run extremely slowly because of the computationally intensive step of checking the reverse translated sequences
- In addition to checking for the primers and cutsites, we also have to check for 'GGGGGG' because it can lead to undesirable structures forming. In addition, we have to check for the reverse complements for all these undesirable sequences.
- We decided on a similarity of 0.8 as the maximum acceptable similarity between the sequence the primer bind to and any other part of the generated sequence. If the sequences are too similar, the primer might mishybridize. We originally had a similarity threshold of 0.6 but that made the program run too slowly, so we decided on a threshold of 0.8.
June 28th
Sequencing:
- the following samples from 6/27 were sent to Genewiz for sequencing:
- PyrF F, R (one sample with PyrF_F, one with PyrF_R)
- rpoz F, R (one sample with rpoz_F, one with rpoz_R)
Lambda red results:
- the colonies on the plates did not look promising, and the ones we chose and grew up in LB+kan did not actually grow. Just to be certain, we choose 18 more colonies: 6 from 37.5µL arabinose 100µL plated, 6 from 3.75µL arabinose 100µL plated, and 6 from 37.5µL arabinose 1.5mL plated. Three from each plate were grown in plain LB and three with kan. We will let it grow in 30˚C, overnight if necessary, and hopefully see bacteria for PCR.
- Assuming this does not work, we prepared more ∆HisB∆PyrF∆rpoZ+pKD46 in two ways: we put 3 colonies in LB+amp from the 6/16 transformation plate, and we streaked a new amp plate from the glycerol stock
- Another possibility is that something is wrong with our lambda red. We designed primers to verify that the pKD46 plasmid is really in the cells.
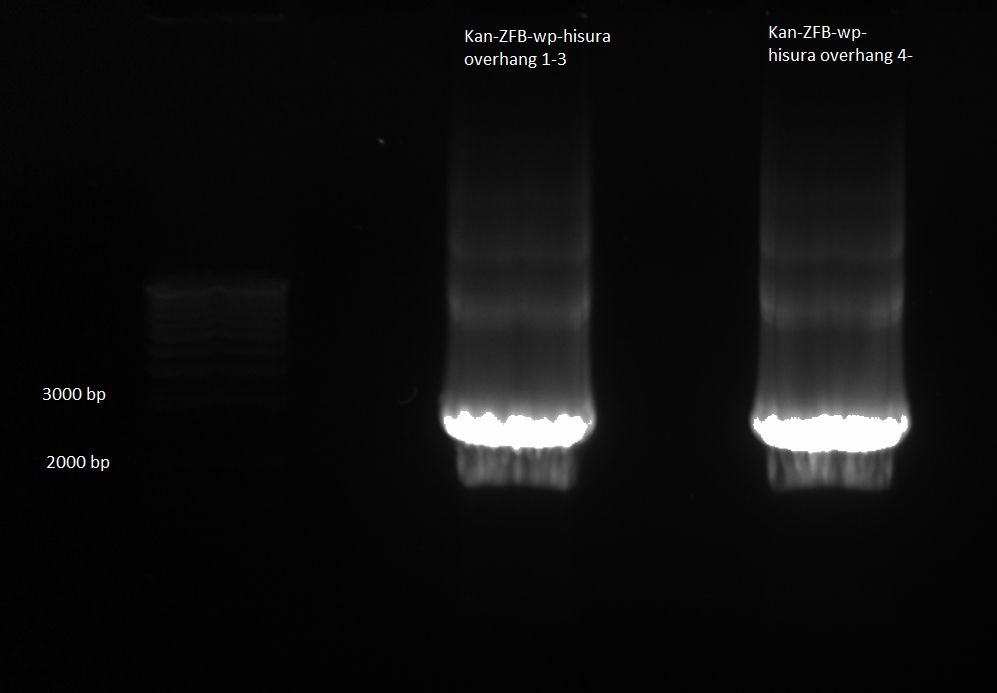
Kan-ZFB-wp-his3-ura3 construct:
- Our last few PCR purifications have given us very low yields, and consequently we have had to use large amounts of our DNA (and the large amounts of buffer salts may also be why our lambda red recombinations have failed). When we tried to amplify our current DNA using the hisura-kan_F and ZFB-wp-hisura_R primers and the Phusion mastermix, it did not work (see 6/23). We will try to gain more product in two ways:
1) Repeat 6/23 PCR but use KAPA mastermix
- the KAPA mix may work better than the Phusion.
- Used KAPA protocol with 1µL of kan-ZFB overlap as template, hisura-kan_F and ZFB-wp-hisura_R primers, 65˚C annealing temp, 90 sec elongation time
- made 2 reactions
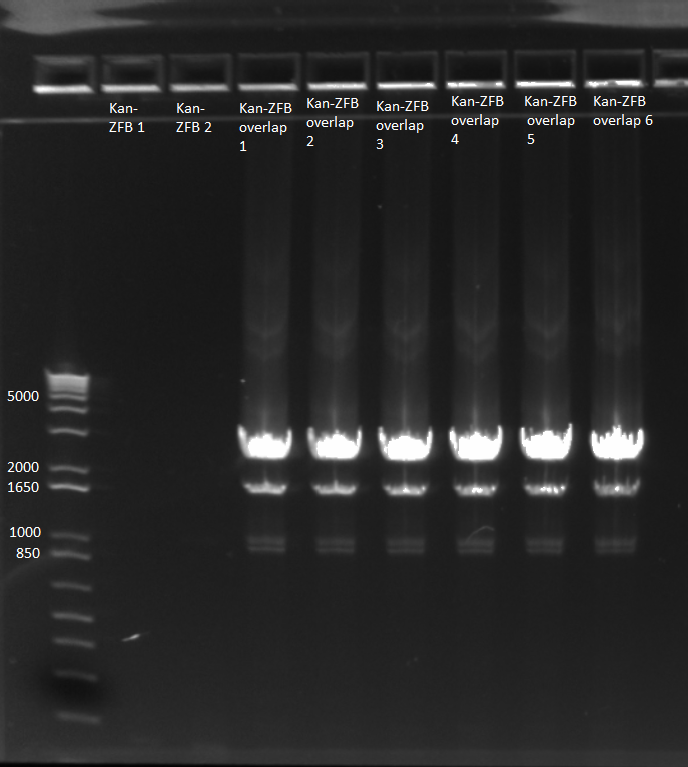
2) Repeat overlap extension PCR (see 6/16) with KAPA mastermix
- used KAPA protocol with 1µL kan cassette and 1µL of ZFB-wp-hisura, 65˚C annealing temp, 90 sec elongation
- 10 cycles without primers; hisura-kan_F and ZFB-wp-hisura_R primers added; 15 more cycles
- made 6 reactions
- E gel to check reactions worked: all 6 overlap PCRs successful, but not the other two reactions.
- combined samples 1-3 and 4-6 and ran on 1% agarose gel for extraction
- used Qiagen gel extraction kit and instructions with the following modifications:
- gel bands were dissolved in 500µL of buffer QG regardless of the gel volume
- gel heated at 50C for 20 min (to make up for reduced amount of buffer QG)
- after melting, 10µL of NaOAC (3M) were added to adjust the pH
- DNA from samples 1-3 were eluted in 20µL of ddH2O; DNA from samples 4-6 were eluted in 20µL of buffer EB
- water sample: 273.4 ng/µL, 260/280=1.92
- EB sample: 136.9 ng/µL, 260/280=2.38
June 28th - Bioinformatics
Attention all Harvard iGEM-ers!!! According to the iGEM Main Page, our preliminary project descriptions and safety proposals are due on July 15. Please see the aforementioned link so we can get this done ASAP- we don't want to miss any deadlines and have all our hard work wasted!
- Finalized our Positive Control Sequences, using Justin's macro to insert the F1 helices into the appropriate zif268 F2 backbone
- Length of chip oligos: 131-140bp (based on Cut Site Design)
- Primers: 20bp (x2= 40bp)
- zif268 F2 backbone + helix= 23aa (x3=69bp; some fingers ~3aa longer)
- Some alternate backbones are longer than zif268 F2 backbone
- Type II binding/cut sites= 11bp on each side (22bp total)
- Standard legnth: 40 + 69 + 22 = 131bp
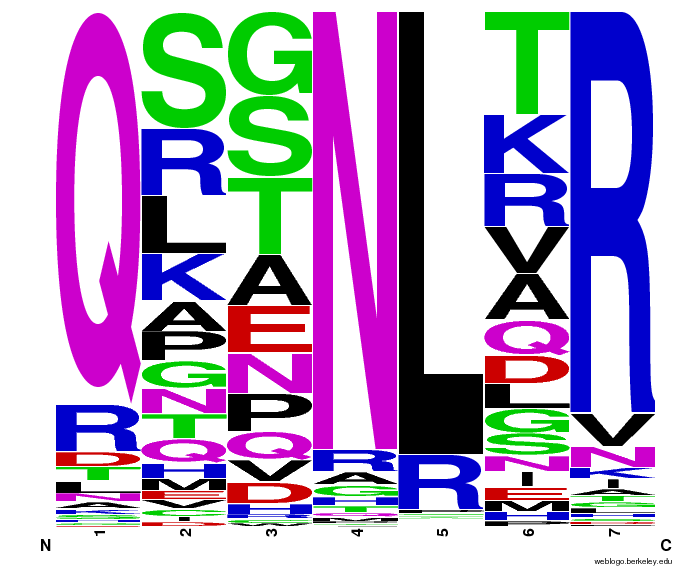
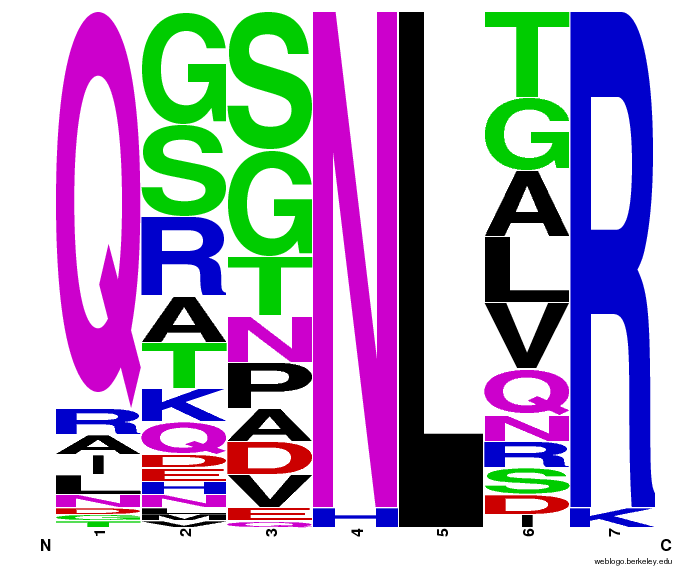
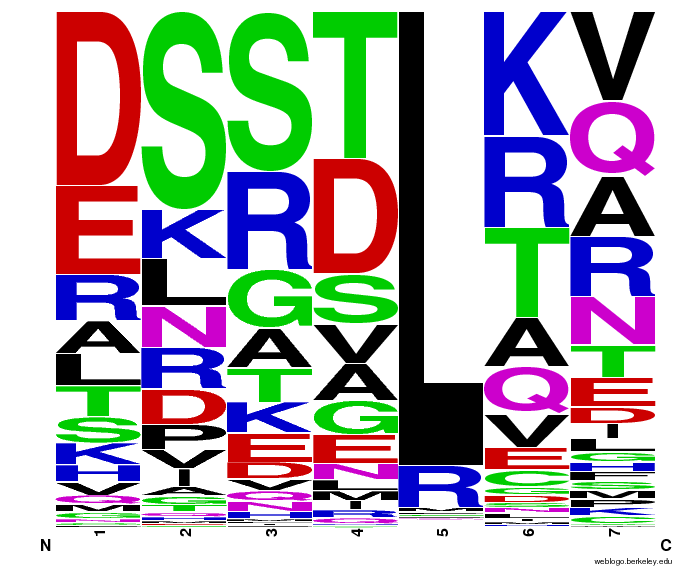
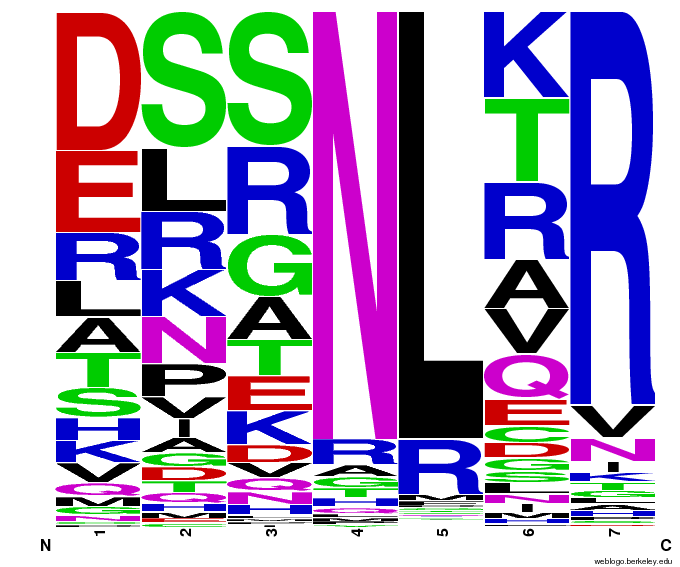
- Use WebLogos as a final visual check of our final generated sequences
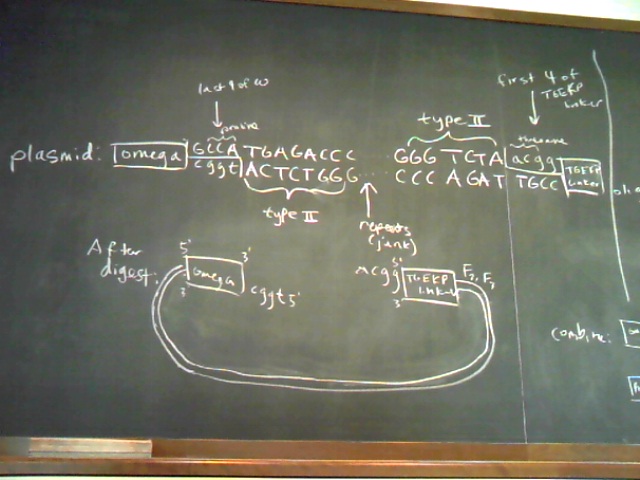
Plasmid and Oligo Design Schematics
Chip-Based Sequence Design Schematic
References
<biblio>
- Kosuri2010 pmid=21113165
</biblio>
Harvard Logo
Running the Generator!
File:HARVFasta total.csv NOTE: LATER GENERATED NEW SEQUENCES. NOT UP TO DATE.
Generated Final Chip Sequences
- We ran the generator once earlier this afternoon, but had to re-run it again due to a typo in the cut sites and the number of sequences we desired for each backbone. Luckily, we caught these errors, and after checking the program once again, we ran it a final time this afternoon.
- It took about 45 minutes for the program to generate and reverse translate the 54900 sequences.
- During this time, we created a function that will re-translate the sequences that the generator output. It compares the original helix with the re-translated helix to make sure that our reverse-translate works properly.
- This step went smoothly, and we verified that the sequences were reverse-translated properly.
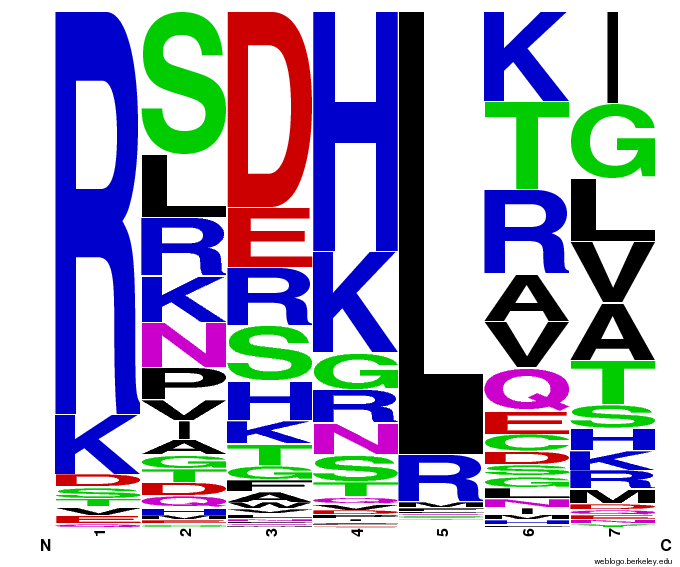
- To make sure that the distributions generated were as expected, we made WebLogos of the helices generated(see below).
- The output file (in the Dropbox: iGem > chip > final chip.csv) originally had the following headers: 'Target', 'Backbone #', 'Helix Sequence', 'Backbone Sequence', 'Nucleotide Sequence of Zinc Finger'
- We wanted to convert this information into FASTA format.
- We wrote a function that converted our original file into fasta format (in the Dropbox: iGem > chip > fasta.csv)
- The file FASTA_total (also linked above) contains the FASTA for all 50000 sequences (including the 100 controls).
- For those curious, the FASTA format just a format that looks like this:
- We wanted to convert this information into FASTA format.
>Header (For us the header is: Target, Backbone #, Helix Sequence, Backbone Sequence)(The header for the controls are: Index Number, 'control') Sequence (In our case, the nucleotide sequence of the zinc fingers)
Generated WebLogos for Final Chip
- FASTA-Formatted Chip Data:
- >NNN(Target Triplet) BB# Helix Seq.
- Nucleotide seq. of ZF
Bioinformatics Candids
File:HARVPrimer Index iGEM 2011
Design of Plate Practice Sequences
While we wait for the chip to come in, we have a number of techniques and protocols that we can practice on beforehand, so that when the chip comes we'll be ready to go to use what they give us. We will be practicing the following techniques:
- Cutting ZF1 out of our oligos
- Inserting ZF1 into the expression plasmid in between the omega subunit and the linker before F2
- Verifying that combination of our F1 from the oligo with the plasmid produces a viable, functional ZF
- Amplifying subpools of oligos for testing
- Inserting the expression plasmids into the E. coli containing our selection genome
- Verifying that our ZF-binding site/GFP expression paradigm works
To this end, we will be ordering a 96-well plate from IDT containing oligos that will simulate the entire tube of oligos that we will receive from Agilent in four weeks. These oligos will consist of the following:
- 6 positive controls (we know which DNA sequences these bind to)
- 3 of them being the F1 fingers of Zif268, OZ052, and OZ123
- 3 of them being ZF F1s derived from CODA.
- 90 generated sequences, picked from a subset of the chip
- These are picked evenly across the 9,150 sequences generated on the cihp for the TGG triplet F1 target from the colorblindness "bottom finger" target, GTG GGA TGG. This particular target was chosen because the F2/F1 is a GNNTNN combo, which might be more likely to get hits from our chip generation sequences.
The primer tag sequences for the 90 generated sequence subset will be the same as they are on the chip (for the sake of explanation, we will refer to them now as P1F and P1R in this paragraph). The positive controls will be flanked immediately by the same primers as the generated subset so that we can amplify everything as one pool altogether should we need to (so this will be P1F and P1R). However, we will also put an additional set of primers outside of the P1F/P1R primers for the positive controls so that we can specifically amplify the positive control subpool, should we want to. These primers will be the same as the primers for the positive control on the chip (which will be called P2F and P2R here).
To recap, on the chip we will have the following oligos :
45750 other oligos for the 5 other target sequences Oligos (TGG set, 9150 total): | P1F | type II binding site | generated F1 | type II binding site | P1R | Oligos (+ control, 100 total): | P2F | type II binding site | control F1 | type II binding site | P2R |
In our test pool of 96 sequences, we will have two types of oligos (note the two pairs of primers around the positive controls):
Oligo (TGG set, 90 total): | P1F | type II binding site | generated F1 | type II binding site | P1R | Oligo (+ control, 6 total): | P2F | P1F | type II binding site | generated F1 | type II binding site | P1R | P2R |
Once we get our test sequences back from IDT, they will come in a 96-well plate with one oligo in each plate. We should make a mixture using some of each well in order to create a tube that contains all 96 sequences. This will simulate the tube that we will receive from Agilent, except instead of 55,000 sequences we will have 96 sequences only in this tube. From here, we can practice using this as a library.
We can pretend that this tube is just 96 generated sequences on the chip, treating the positive controls as if they were also generated sequences (we only include them in the 96 to ensure that we will indeed get a "hit" from this practice screening). Thus, we can just use the P1F/P1R primer set to amplify all of them in order to use them for the subsequent steps.
These subsequent steps will be those that were outlined above, namely cutting out the F1 sequence from each oligo, ligating this F1 into our expression plasmid, putting the expression plasmid into our selection strain, observing colonies which get infused with ZFs that bind to our target site (the "hits"), and sequencing the colonies that get hits to determine which ZF they are expressing.
We will be repeating these exact same steps once we get the chip, so if we can perfect our protocols with these practice sequences, we should be golden when the chip comes in.June 29th
Our first day with everyone in the wet lab!
PyrF and rpoZ sequencing:
- For some reason, Genewiz said that sequencing failed due to "no priming." We will redo the PCR and send the products in again today.
Lambda red recombination and MAGE:
- The cultures made from the kan plates from our earlier attempt at lambda red did (in some cases) grow, including one colony in kan from 3.75µL arabinose, 100µL plated.
- PCR of liquid culture: used 1µL of culture diluted 1:20 as well as saturated culture of ∆hisB∆pyrF∆rpoZ+pKD46 (diluted 1:20) as a 1529620 locus wild-type control
- primers (from Vatsan): 1529481-f, 1529806-r
- KAPA mastermix and procedure with 56˚C annealing and 90 seconds elongation
- E Gel of product: both wild-type and the sample hopefully containing the insert had the same short band of around 350bp--the recombination was unsuccessful
- PCR of liquid culture: used 1µL of culture diluted 1:20 as well as saturated culture of ∆hisB∆pyrF∆rpoZ+pKD46 (diluted 1:20) as a 1529620 locus wild-type control
- Used overnight saturated culture to reinoculate; once close to mid-log, culture split into 2 1.5mL amounts and 37.5µL arabinose added to each
- same procedure as previously described, but with about 300ng kan-ZFB construct and a 2.5µM final concentration of HisBNuke3 (12.5µL of 10µM stock)
- electroporate with 1.8 kV, about 5
- recover 3 hrs
- kan-ZFB insertion colonies plated on kan, MAGE on amp
PCR Preparation
- Lambda Red- Selection strain glycerol stock: 1/100 dilution, 2 uL stock with 198 uL ddH2O
- Spec (Colony)- Touch 1 colony with pipet tip, add and mix with pipet in 20 uL ddH2O, then vortex
PCRs
- PKD46 (Lambda Red)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 1 uL
- ddH2O- 10 uL
- (25 uL total)
- Spec (Colony)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 1 uL
- ddH2O- 10 uL
- (25 uL total)
- Spec (Miniprep)
- Kapa Mix 2x- 12.5 uL
- Primer_F- 0.75 uL
- Primer_R- 0.75 uL
- Template 2 uL
- ddH2O- 9 uL
- (25 uL total)
Expression Plasmid Design in silico
Today, we designed our expression plasmids in SeqBuilder. This included plasmids for our 6 target sequences, and 3 positive controls (9 in total). These positive controls were the following:
| Index | Nucleotide Sequence (5\'-3\') | Helices (F3 to F1) | Notes |
| 16 | GAA GGG AAC | QDGNLGR RREHLVR HRTNLIA | Very similar to one of our target sequences (CB top), which is GAA GGG ACC |
| 55 | GGA GTG GTG | QTTHLSR DHSSLKR RNFILQR | Very similar to a target sequence (FH top), which is GGA GTG CTG |
| 77 | TGT GAA TAG | RRRNLQI QQTNLTR QPHGLTA | Out of ze air |
Each of our expression plasmids contained:
- Omega subunit
- Omega/F1 linker (taken from paper that Dan and Noah emailed us), http://nar.oxfordjournals.org/content/36/8/2547.short
- type II binding sites
- gap between type II binding sites that contains XbaI restriction enzyme site (which is not present anywhere else in the entire expression plasmid)
- F1/F2 TGEKP linker
- F2 for a specific target sequence
- F2/F3 TGEKP linker
- F3 for a specific target sequence
- TAA stop codon immediately after F3
All of this is on our spec-resistance-containing plasmid. The above construct replaced the GFP which was present previously on this plasmid.
Tomorrow we will begin design of our primers from these SeqBuilder files.June 30th
Lambda Red, Backbone, and Sequencing PCR
- Gel run on the presence of a Lambda Red protein in the pKD46 plasmid showed that it is indeed present, so our recombination failures have not been due to an incorrect plasmid.
- Gel run on the backbone of pZE21G plasmid was success and took us one step closer to obtaining all parts necessary for the three part assembly
- Gel run on the pyrF and rpoZ was success
- Therefore we sent the PCR products and primers to GENEWIZ for sequencing again
pZE21G backbone:
- Since last night's PCR was successful, we will redo it with a few protocol adaptations to get a cleaner product and to increase our yield when we purify
- KAPA mastermix and protocol: primers HindIII-F and KpnI-R
- template: pZE21G miniprepped plasmid, 1µL
- 2 min elongation time, 30 cycles
- 2 samples at 55˚C annealing, 2 samples at 60˚C
Lambda red and MAGE:
- Yesterday's prep produced tiny colonies on the MAGE plates and (so far) none on the kan-ZFB plates. Just in case it didn't work, we will redo the lambda red using even more DNA and perform a second round of MAGE using culture from yesterday that was not plated.
- same procedure, but with the following changes:
- 5µL kan-ZFB (about 1 mg)
- recover 3 hrs
- kan-ZFB: plate 100µL and 2 mL on kan plates
- MAGE: plate 1µL and 10µL on amp plates
- To see if the colonies on the MAGE plate knocked out HisB, we chose 24 colonies, resuspended them in water, and put half the cells in LB (complete media) and half in NM media (does not have histidine). 96 well plate, 150µL media, grown overnight at 30˚C.
ZF Expression Plasmid Ultramer and Primer Design
Today, we designed primers ZF_073 through ZF_085 as listed in the iGEM Primer Index spreadsheet. These were basically two sets of primers: the primers to clone out the omega subunit and linker, and the ultramers that would construct the last part of the linker along with the type II binding sites and F2/F3 fingers. One should refer to the primer list for the sequences.
Note: the annealing sequence for the ultramer overlap contained a 72 degree melting temp hairpin. To get around this, I changed one of the codons in the F2 backbone. The F2 backbone begins with "FQCRIC", and so I changed the codon for the arginine (R) from CGC to CGT, which resolved the hairpin problem.
June 30th - Bioinformatics
Updated Primer list and FASTA formatting
We ran into a small hiccup, when we were informed that we had forgotten to reverse translate the reverse primer sequences that were being appended to the generated sequence. This is because the primer sequences we were given were the sequences for the actual primers, rather than sequences to which the primers would bind. Luckily, we caught this error! We did have the re-run the generator because we had to make sure that our generated sequences did not contain the new primers.
- Here is the updated primer list:
This is the set of final target sequences with assigned forward and reverse primers (tags for PCR):
| Disease | Target Sequence | Forward Primer (5'-3' NOT REVERSE COMPLEMENT) | Reverse Primer (5'-3' REVERSE COMPLEMENT) |
| Colorblindness | GCT GGC TGG | ATATAGATGCCGTCCTAGCG | TGGGCACAGGAAAGATACTT |
| Colorblindness | GCG GTA ACC | CCCTTTAATCAGATGCGTCG | GGTCGCCCTTATTACTACCA |
| Familial Hypercholesterolemia | GGC TGA GAC | TTGGTCATGTGCTTTTCGTT | TCTGAGTATCCGATACCCCT |
| Familial Hypercholesterolemia | GGA GTC CTG | GGGTGGGTAAATGGTAATGC | GCTATATCCGGGGAATCGAT |
| Myc-gene Cancer | GGC TGA CTC | TCCGACGGGGAGTATATACT | TTGGCCTGAAGCAGTTAGTA |
| Myc-gene Cancer | GGC TGG AAA | CATGTTTAGGAACGCTACCG | GGGAGGGAACGGAGATTATT |
| Controls | n/a | GTACATGAAACGATGGACGG | CGCTGAGGAGACTATACCAG |
There was also a small error in the FASTA formatting. There are not supposed to be any spaces in the header, so the spaces were replaced with underscores.
- Example:
>1_controlGTACATGAAACGATGGACGGGGTCTCAGCCATTCCAATGTCGTATCTGTATGCGTAATTTTTCACGCAAACACCATTTGGGTCGTCATATCCGTACGCACACGGTGAGACCCGCTGAGGAGACTATACCAG
 "
"