Team:Harvard/Template:NotebookDataJuly2
From 2011.igem.org
(Difference between revisions)
| Line 20: | Line 20: | ||
**samples prepared for sequencing tomorrow | **samples prepared for sequencing tomorrow | ||
| - | [[File: | + | [[File: HARV2011.07.07.hisBseq labeled.png|thumb|none|MAGE 2 samples for sequencing 7/7/11]] |
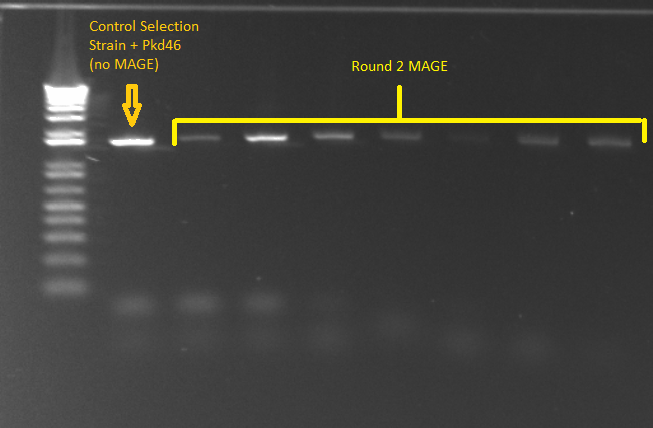
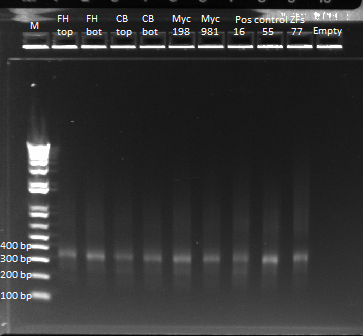
*Kan-ZFB insert check PCR results: | *Kan-ZFB insert check PCR results: | ||
| Line 26: | Line 26: | ||
**Results: some of the reactions, strangely, did not work, but the ones that did showed a short band and thus did not have the insert (see below) | **Results: some of the reactions, strangely, did not work, but the ones that did showed a short band and thus did not have the insert (see below) | ||
| - | [[File: | + | [[File: HARV2011.07.07,kanZFBinsert_check1 labeled.png|thumb|none|1529620 locus PCR 7/7/11]] |
| - | [[File: | + | [[File: HARV2011.07.07,kanZFBinsert_check2 labeled.png|thumb|none|1529620 locus PCR 7/7/11]] |
*HisBNuke3 MAGE round 3: | *HisBNuke3 MAGE round 3: | ||
| Line 49: | Line 49: | ||
| - | [[File: | + | [[File:HARVKAN-ZFB-WP.png|200px|thumb|none|Kan-ZFB-WP]] |
===Web Design=== | ===Web Design=== | ||
| Line 98: | Line 98: | ||
*Results: PCR only produced a smaller side product. This is strange since we also ran a gel of all our different kan-ZFB-hisura purifications and they all run at the correct size (see images below). | *Results: PCR only produced a smaller side product. This is strange since we also ran a gel of all our different kan-ZFB-hisura purifications and they all run at the correct size (see images below). | ||
| - | [[File: | + | [[File: HARV2011.07.08.ECnr2MAGEround2-21to24delrpoZ&thioKAN-ZFB labeled.png|thumb|none|Thio kan-ZFB 7/8/11]] |
| - | [[File: | + | [[File: HARV2011.07.08.kanzfbDNA(labeled).png|thumb|none|kan-ZFB insert purifications]] |
===Team TolC=== | ===Team TolC=== | ||
| Line 149: | Line 149: | ||
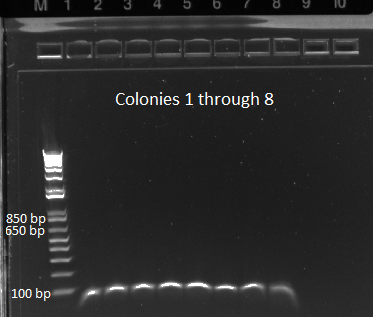
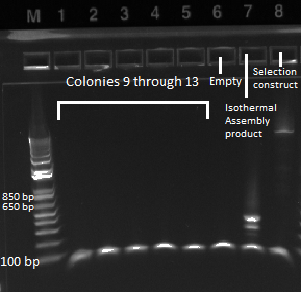
We ran an agarose gel to check the bands: | We ran an agarose gel to check the bands: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011_07_10_transformation_colony_pcr_lane_1-8.png|thumb|Lanes 1-8]] |
| - | |[[File: | + | |[[File:HARV2011.07.10_transformation_colony_pcr_lane_9-16.png|thumb|Lanes 9-16]] |
|} | |} | ||
| Line 221: | Line 221: | ||
We ran a 1% 100 ml. agarose gel at 120V to check the success of the PCR. | We ran a 1% 100 ml. agarose gel at 120V to check the success of the PCR. | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV7.11.11.omegazif_transformed_annotated.png|thumb|Gel of our colonies, miniprep-ed colonies, and controls to check for w+zif268 insertion]] |
|} | |} | ||
| Line 228: | Line 228: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV7.11.11.omegazif_transformed_egel_annotated.png|thumb|Gel to ensure our bands were the correct size (709 bp)]] |
|} | |} | ||
| Line 240: | Line 240: | ||
**KAPA protocol using M13_F and M13_R primers (anneal to the vector), 50˚C annealing, 2min elongation | **KAPA protocol using M13_F and M13_R primers (anneal to the vector), 50˚C annealing, 2min elongation | ||
**E gel results: lots of bands but rather messy and not exactly what we'd expect. We'll try to verify the presence of the plasmid in a few other ways. | **E gel results: lots of bands but rather messy and not exactly what we'd expect. We'll try to verify the presence of the plasmid in a few other ways. | ||
| - | [[File: | + | [[File: HARVPSR01 test1(labeled).png|thumb|none|pSR01 colony PCR 7/11/11]] |
*Sequencing: | *Sequencing: | ||
**We sent out the PCR samples with M13_R to Genewiz | **We sent out the PCR samples with M13_R to Genewiz | ||
| Line 250: | Line 250: | ||
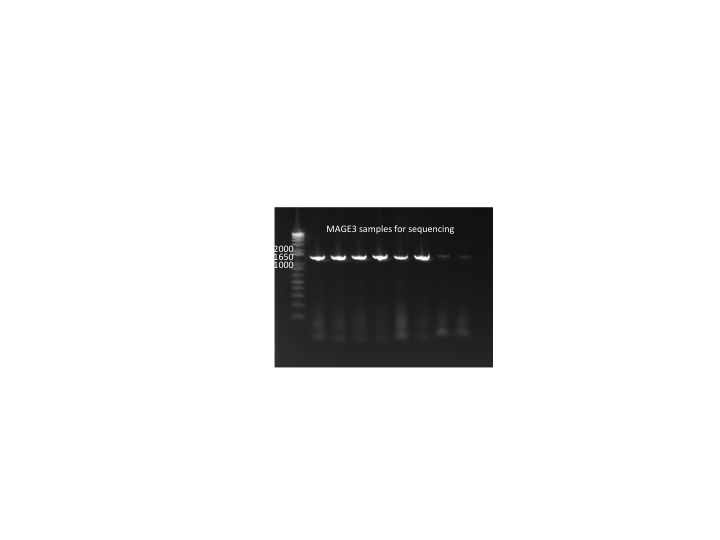
*In case pSR01 does not rescue the selection strain's growth phenotype, we will PCR 96 colonies from the MAGE3 plates and send them out for sequencing tomorrow. | *In case pSR01 does not rescue the selection strain's growth phenotype, we will PCR 96 colonies from the MAGE3 plates and send them out for sequencing tomorrow. | ||
**Used 9µL KAPA, His_F and His_R primers, 3µL suspended colony (out of 7--the remaining 4 grown in LB+amp) | **Used 9µL KAPA, His_F and His_R primers, 3µL suspended colony (out of 7--the remaining 4 grown in LB+amp) | ||
| - | [[File: | + | [[File: HARV2011.07.11.MAGE 3 96 A1-H1.png|thumb|none|MAGE3 colonies for sequencing 7/11/11]] |
'''Thio kanZFB:''' | '''Thio kanZFB:''' | ||
*We retried the PCR to put modified nucleotides on the ends of the kan-ZFB-wp-hisura insert using the same procedure as 7/8/11 | *We retried the PCR to put modified nucleotides on the ends of the kan-ZFB-wp-hisura insert using the same procedure as 7/8/11 | ||
**E gel: reactions didn't work | **E gel: reactions didn't work | ||
| - | [[File: | + | [[File: HARV2011.07.11.thiokanZFB(labeled).png|thumb|none|thio kan-ZFB PCR 7/11/11]] |
*Tried again with a longer elongation time (3 min) and let run overnight. | *Tried again with a longer elongation time (3 min) and let run overnight. | ||
| Line 276: | Line 276: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.12_nine_ultramers_product_annotated.png|thumb|Gel of our product from the ultramer PCR. Note that the first lane's PCR was done in a single tube, while all the others were done in a strip.]] |
|} | |} | ||
| Line 282: | Line 282: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.12_nine_ultramers_product_try2_annotated.png|thumb|Gel of our product after PCR purification ]] |
|} | |} | ||
| Line 294: | Line 294: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.12_omega+linker_and_crossjunction_omgzif-spec_plasmid_annotated.png|thumb|'''Lane 3''': Gel of our PCR to check for the cross-junction in colony 9.]] |
|} | |} | ||
| Line 303: | Line 303: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.12_omega+linker_and_crossjunction_omgzif-spec_plasmid_annotated.png|thumb|'''Lanes 1 and 2''': Gel of our PCR to get omega subunit.]] |
|} | |} | ||
| Line 312: | Line 312: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.12_OZ052_and_OZ123_ultramers_(last_two_lanes)_annotated.png|thumb|'''Last two lanes''': OZ052 and OZ123 products, respectively]] |
|} | |} | ||
| Line 324: | Line 324: | ||
**In case this mutation is correct but lambda red in the selection strain still eludes us, we designed MAGE oligos for use in EcNR2 to either delete the two aa or put 3 stop codons after the HPase region. | **In case this mutation is correct but lambda red in the selection strain still eludes us, we designed MAGE oligos for use in EcNR2 to either delete the two aa or put 3 stop codons after the HPase region. | ||
*culture PCR results: cultures 3-4 had a strong band (and 2 and 5 weak ones) around 850-1000bp. It is interesting how different these results are from yesterday's colony PCR gel. | *culture PCR results: cultures 3-4 had a strong band (and 2 and 5 weak ones) around 850-1000bp. It is interesting how different these results are from yesterday's colony PCR gel. | ||
| - | [[File: | + | [[File: HARV2011.07.12.pSR01 with phospho primers(labeled).png|thumb|none|pSR01 culture and thio kan-ZFB PCRs 7/12/11]] |
*Sequencing results: The sequences sent from Genewiz aligned well with the pSR01 plasmid sequence and covered pretty much all of URA3. This implies that the cells did indeed take up our plasmid. | *Sequencing results: The sequences sent from Genewiz aligned well with the pSR01 plasmid sequence and covered pretty much all of URA3. This implies that the cells did indeed take up our plasmid. | ||
*Growth phenotype: | *Growth phenotype: | ||
| Line 371: | Line 371: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.13_ultramers_annotated.png|thumb|Gel of our ultramer touchdown PCR product]] |
|} | |} | ||
| Line 408: | Line 408: | ||
***KAPA, 1529620-flanking primers, 90 sec elongation and 56 anneal, 25 cycles | ***KAPA, 1529620-flanking primers, 90 sec elongation and 56 anneal, 25 cycles | ||
***Results: 300bp band--the streaks on the plate are bacteria, but they do not have the insert. | ***Results: 300bp band--the streaks on the plate are bacteria, but they do not have the insert. | ||
| - | [[File: | + | [[File: HARV2011.07.13 Lambda red streaky plate(labeled).png|thumb|none|1529620 locus PCR of EcNR2 lambda red 7/13/11]] |
*We also retried the thio kan-ZFB PCR using some new primer aliquots. Once again it failed. | *We also retried the thio kan-ZFB PCR using some new primer aliquots. Once again it failed. | ||
| - | [[File: | + | [[File: HARV2011.07.13.Phospho thio 4(labeled).png|thumb|none|Thio kan-ZFB PCR 7/13/11]] |
*Because this PCR keeps failing, we're going to try a different way and do two separate and shorter PCRs to be joined later in an overlap PCR: | *Because this PCR keeps failing, we're going to try a different way and do two separate and shorter PCRs to be joined later in an overlap PCR: | ||
**PCR of kan cassette: kan cassette as template (diluted 6/16 aliquot), kan F phosphothio and kan R primers, KAPA, 56˚C anneal and 90 sec elongation | **PCR of kan cassette: kan cassette as template (diluted 6/16 aliquot), kan F phosphothio and kan R primers, KAPA, 56˚C anneal and 90 sec elongation | ||
| Line 429: | Line 429: | ||
***10.7 ng/ | ***10.7 ng/ | ||
| - | [[File: | + | [[File: HARV2011.07.14.TolCinsertionconstruct(labeled).png|thumb|none|KAN-ZFB-wp construct with tolC homology 7/13/11]] |
===Team Web Design=== | ===Team Web Design=== | ||
| Line 446: | Line 446: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.14_9_ultramer_isotherm_asm_rxn_with_junction_primers_annotated.png|thumb|1.2% E-gel of our ultramer isothermal assembly reaction product]] |
|} | |} | ||
| Line 457: | Line 457: | ||
{| | {| | ||
| - | |[[File: | + | |[[File:HARV2011.7.14_ultramers.pcr1_touchdown_pcr_iso_assm_annotated.png|thumb|Products ran in an agarose gel. Note that wells 78-80 on the left side of the gel are empty due to sample mysteriously floating out of the well.]] |
|} | |} | ||
| Line 469: | Line 469: | ||
'''thio kan and thio ZFB-wp-hisura PCR:''' | '''thio kan and thio ZFB-wp-hisura PCR:''' | ||
*Ran overnight PCR of kan cassette and ZFB-wp-hisura on E gel: looks great! | *Ran overnight PCR of kan cassette and ZFB-wp-hisura on E gel: looks great! | ||
| - | [[File: | + | [[File: HARV2011.07.14.thiokan+thioZFBhisura(labeled).png|thumb|none|thio kan and thio ZFB-wp-hisura PCR 7/14/11]] |
*PCR purification with Qiagen kit: | *PCR purification with Qiagen kit: | ||
**added an additional 300µL buffer PB to adjust the pH | **added an additional 300µL buffer PB to adjust the pH | ||
| Line 479: | Line 479: | ||
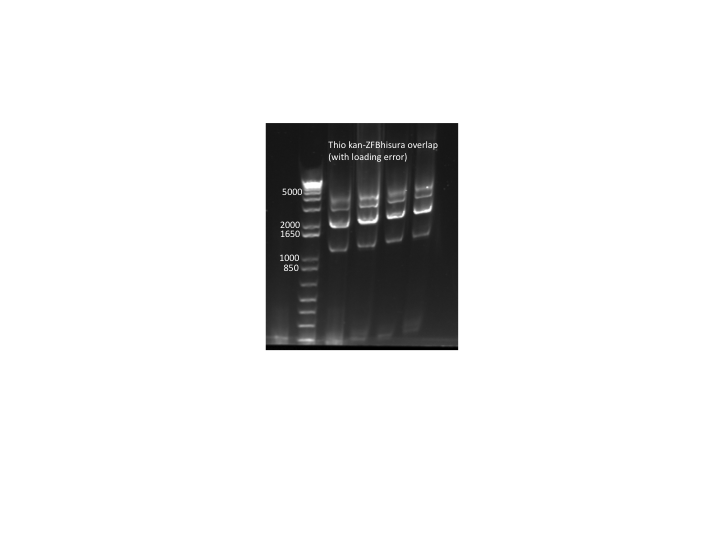
**primers (kan f phosphothio and ura3 r phosphothio) added after 10 cycles | **primers (kan f phosphothio and ura3 r phosphothio) added after 10 cycles | ||
**Results: PCR worked but with tons of side products so we will try to optimize the procedure. Expected product = 3Kb, notable side products = 2Kb, 4Kb. N.B. The samples were accidentally loaded while the gel was running so they are staggered on the gel. | **Results: PCR worked but with tons of side products so we will try to optimize the procedure. Expected product = 3Kb, notable side products = 2Kb, 4Kb. N.B. The samples were accidentally loaded while the gel was running so they are staggered on the gel. | ||
| - | [[File: | + | [[File: HARV2011.07.14.thio kanZFBhisura overlap(labeled).png|thumb|none|thio kan-ZFB-wp-hisura overlap PCR (with loading error)]] |
*Gel stab PCR | *Gel stab PCR | ||
** DNA from the 3Kb e-gel band were removed using the stabbing technique and placed in 4 respective reaction tubes (diluted in 20µL water) | ** DNA from the 3Kb e-gel band were removed using the stabbing technique and placed in 4 respective reaction tubes (diluted in 20µL water) | ||
| Line 496: | Line 496: | ||
**Primers were TolC_seq_F and TolC_seq_R. | **Primers were TolC_seq_F and TolC_seq_R. | ||
***Without insertion these two primers will create 449 bp sequence, and with insertion length will be 1650 bp | ***Without insertion these two primers will create 449 bp sequence, and with insertion length will be 1650 bp | ||
| - | [[File: | + | [[File: HARV2011.07.06.TolCinsertionconstruct+negativecontrol(labeled).png|thumb|none|TolC insert check 1 7/14/11]] |
**Some of the original culture after recombineering, 1ml solution spun down, and resuspended in 100μl of LB, and plated on new KAN plates. To check, one of the old KAN plates was plated the same way and put in at 2:30pm. (These plates will be checked tomorrow morning.) | **Some of the original culture after recombineering, 1ml solution spun down, and resuspended in 100μl of LB, and plated on new KAN plates. To check, one of the old KAN plates was plated the same way and put in at 2:30pm. (These plates will be checked tomorrow morning.) | ||
*PCR done of a few of the rpoZ MAGE with stop codon insertions. Oligo name: rpoZ_MAGE_KO (Noah's oligo) (Noah's plates, after two cycles of MAGE, on both ECNR2 and ECNR2ΔTolC). The side product in the gel could be due to a low annealing temperature of 53°C, so we should do a gradient PCR (e.g.55-66°C) to get the most product the next time. </div> | *PCR done of a few of the rpoZ MAGE with stop codon insertions. Oligo name: rpoZ_MAGE_KO (Noah's oligo) (Noah's plates, after two cycles of MAGE, on both ECNR2 and ECNR2ΔTolC). The side product in the gel could be due to a low annealing temperature of 53°C, so we should do a gradient PCR (e.g.55-66°C) to get the most product the next time. </div> | ||
 "
"