Team:Caltech/Week 7
From 2011.igem.org
| Line 206: | Line 206: | ||
HPLC reveals shift in the chemical structure of BPA after incubation with p450's | HPLC reveals shift in the chemical structure of BPA after incubation with p450's | ||
==July 30== | ==July 30== | ||
| - | Take out plates from fosmid titering | + | Take out plates from fosmid titering<br/> |
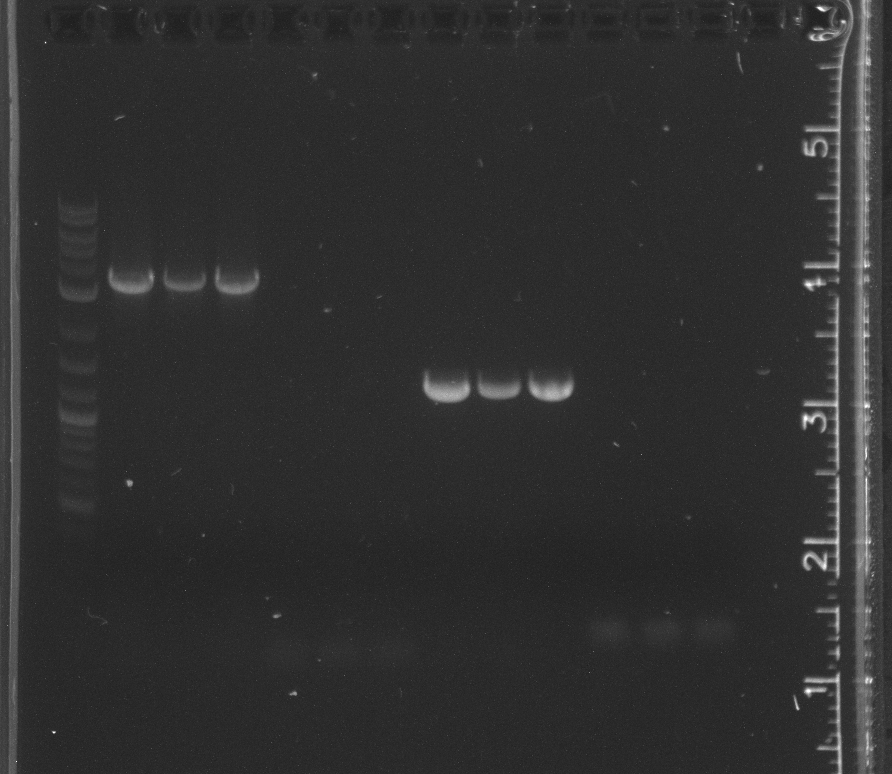
| + | Ran gels of PCR of pNT002 insert, pNT003 insert and pSB3K3 from yesterday.<br/> | ||
===Results=== | ===Results=== | ||
| - | Two colonies on both 9mix and 10-2mix fosmid 1x dilutions | + | <p>Two colonies on both 9mix and 10-2mix fosmid 1x dilutions</p> |
| + | <gallery> | ||
| + | File:7-30pcrgel1.jpg|lane 1 NEB 2-log ladder, 2-4 pNT002 insert, 5-7 pNT003 insert, 8-10 pSB3K3 | ||
| + | File:7-30pcrgel2.jpg|PCR of pNT003 insert with increasing DMSO:lane 1 NEB 2-log ladder, 2-3 0% DMSO, 4-5 0.5%, 6-7 1%, 8-9 2%, 10-11 3% | ||
| + | </gallery> | ||
| + | pSB3K3 was not amplified. The DMSO does not to appear to have decreased the 400 bp band or increased the correct 2 kb band.<br/> | ||
}} | }} | ||
Revision as of 05:10, 1 August 2011
|
Project |
July 24Started overnight cultures of pSB3K3 July 25PCR'd parts for pNT002: R0040, K123001, B0014, pSB4A5 ResultsDecided not to use the pNT003 PCR'd insert, as the band of the correct length was much fainter than a lower length band. Chose not to do Gibson assembly of the positive GFP control, despite not having any green colonies last week, instead focusing on assembling pNT002 and pNT003. PCR concentrations
PSB3K3 miniprep concentrations
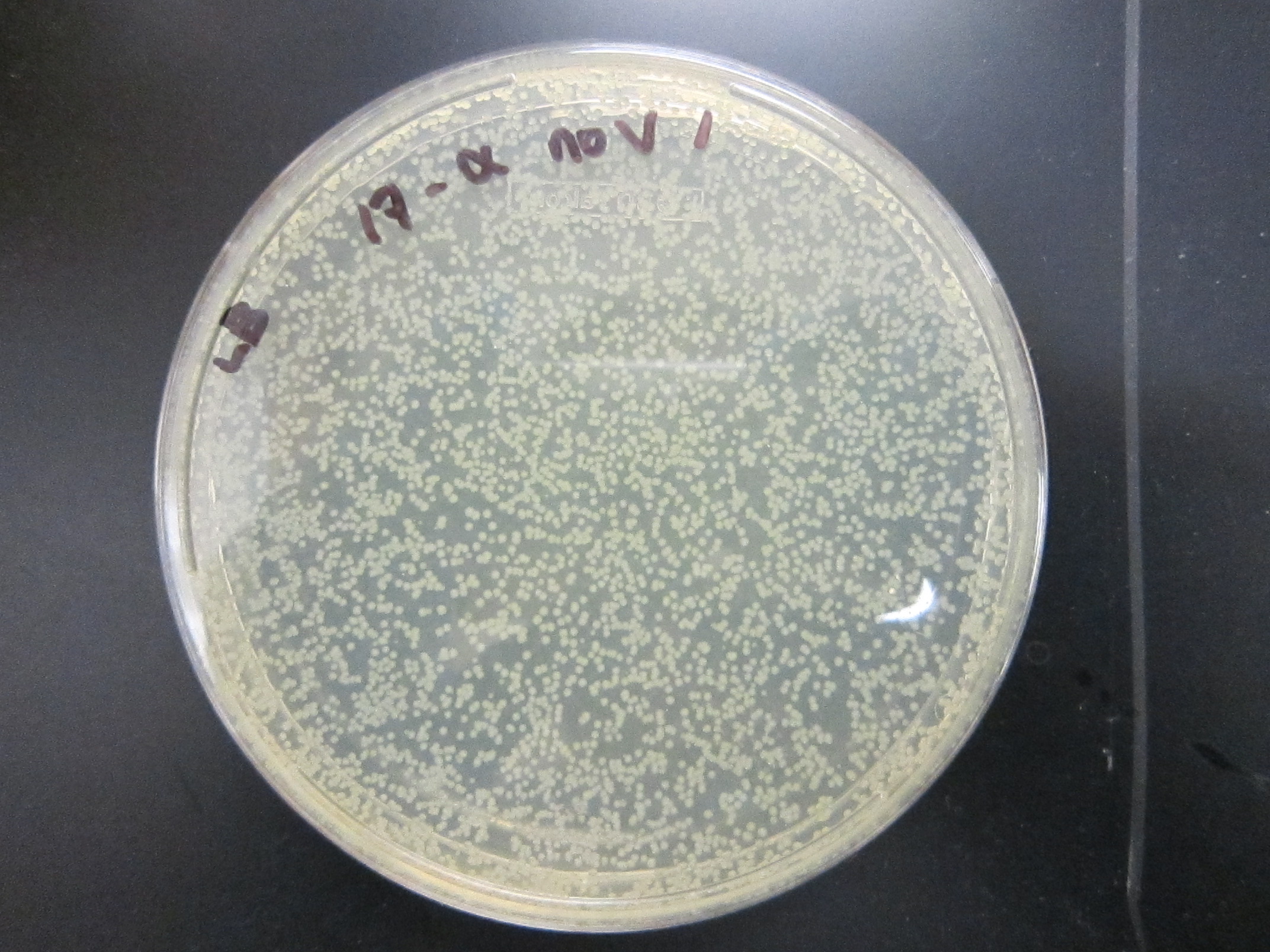
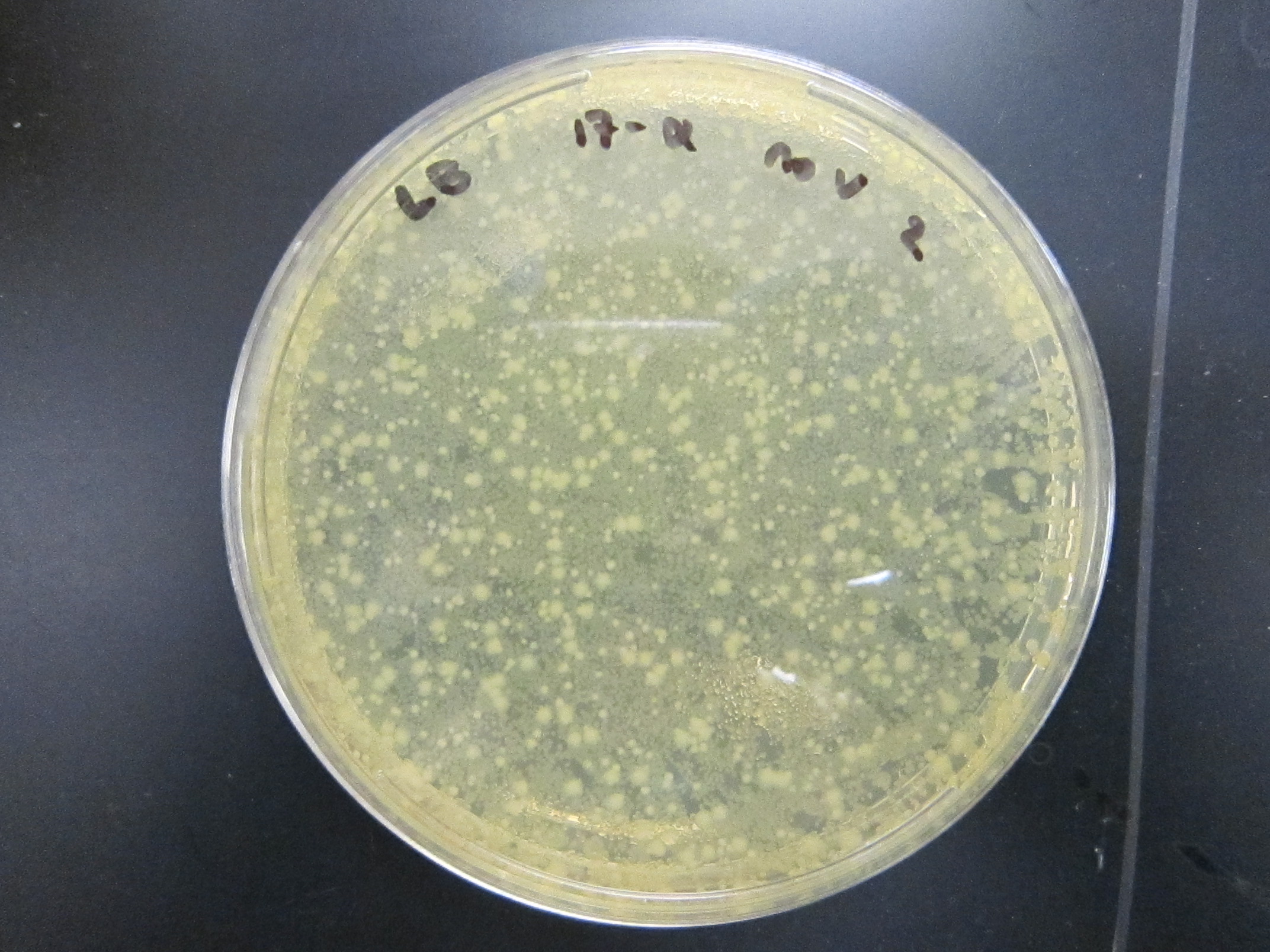
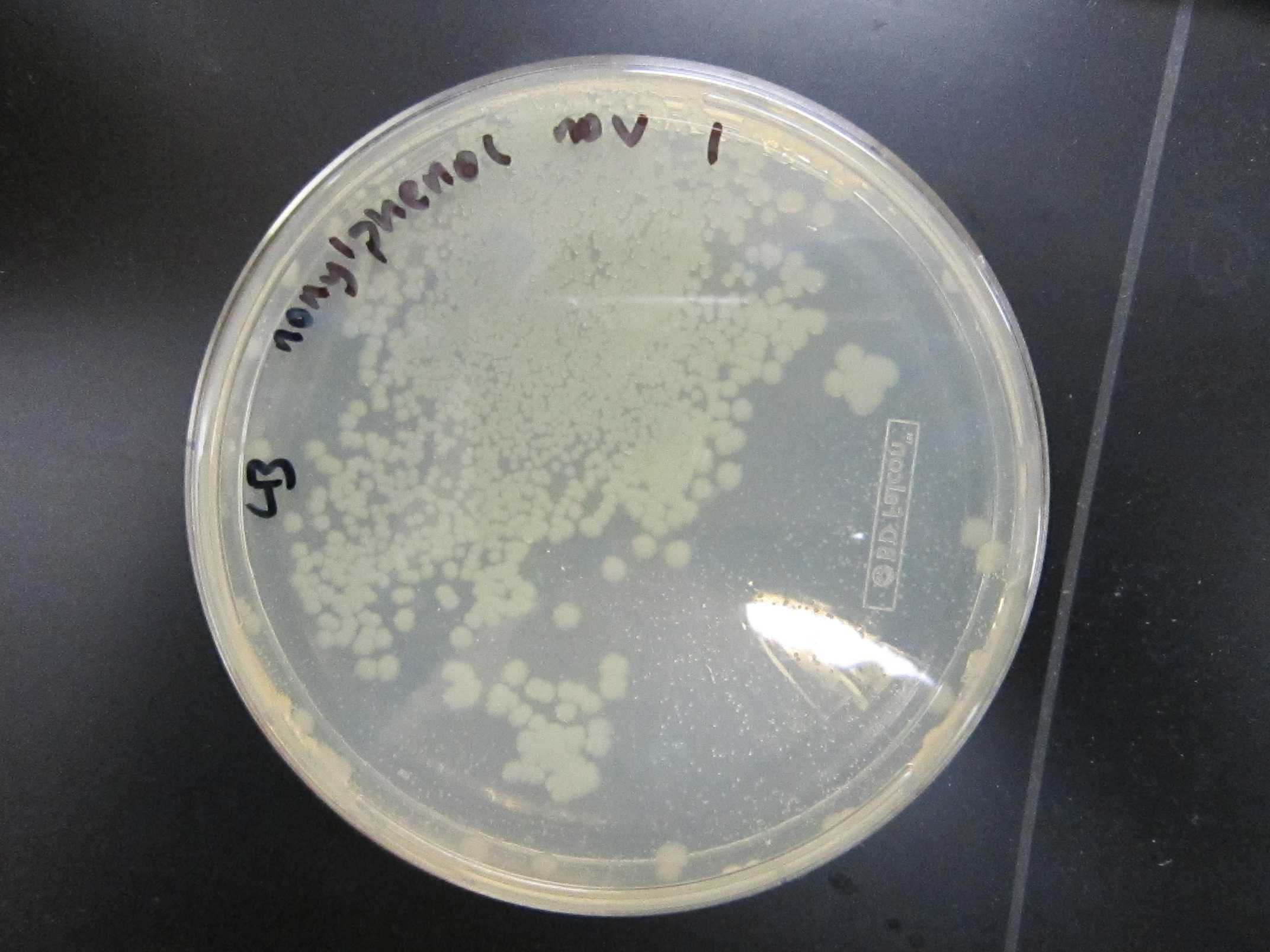
July 26Packaging of 9mix and (10mix + 10-2) ligations Results
The gel gave no bands in any lane, with or without the restriction digest. pNT003 + without the Spe1 digest had a faint smear, the other lanes had nothing. Chose to transform anyway
July 27Grew up cells for titering of packaged fosmids; reached OD600 of 0.974 ResultsThe 16s PCR from yesterday, shown in the gel today, had a band in the first lane of negative controls, indicating contamination. The pSB3K3 amplification failed. The bands indicated a much lower length than expected. After doing an alignment in Geneious, it appears our pSB3N5 primers we ordered for pSB3C5, which we are no longer using because of our competent cell strain, do not work with earlier versions of the 3 origin plasmids. Both the negative control and experimental of PCR purified Gisbson assembly from yesterday of pNT003 had 0 colonies. The transformation with 1 ul pUC 19 had no colonies.
BPA solutions fail to form uniform layers when poured on top of agar, but form a uniform layer when poured directly onto a clean plate. July 28Plate packaged fosmids to obtain titer ResultsYesterday's 16s PCR failed, as there were no bands in control or experimental lanes. The vector was amplified and was the correct length. We redid it, but again there were no bands in the control or experimental lanes. Enrichment cultures plated in minimal media with chemical layer, to be checked in a few days.
July 29Attempted to PCR pNT002 and pNT003 inserts from the purified gel extractions of July 27, using the same primers. ResultsHPLC reveals shift in the chemical structure of BPA after incubation with p450's July 30Take out plates from fosmid titering ResultsTwo colonies on both 9mix and 10-2mix fosmid 1x dilutions pSB3K3 was not amplified. The DMSO does not to appear to have decreased the 400 bp band or increased the correct 2 kb band.
|
 "
"