Team:NTNU Trondheim/Journal
From 2011.igem.org
m (→Tuesday 26/7) |
m (→Wednesday 27/7) |
||
| Line 1,245: | Line 1,245: | ||
== Wednesday 27/7 == | == Wednesday 27/7 == | ||
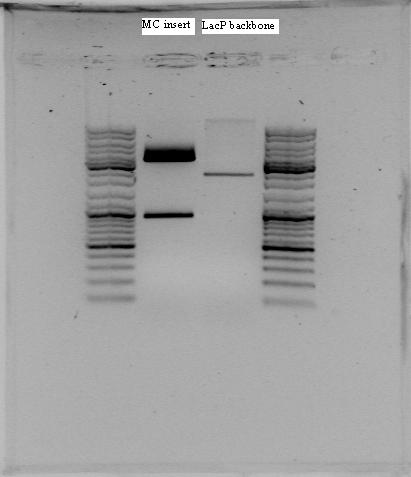
| - | The PCR-amplified RelA was digested with EcoRI and SpeI and purified. Concentration of the sample isolated from 0,5 µl template was 5,9 ng/µl while the one isolated from 5 µl was 4,3 ng/µl. The RelA insert cut with | + | RelA was amplified using PCR. The PCR product of our first amplification of RelA served as template DNA. Since the concentration of the template DNA was low, we amplified two different samples of it. In the first sample, we added 0.5 µl template DNA and 41.5 µl dH<sub>2</sub>O, and in the second sample, we used 5 µl of template DNA and 37 µl of dH<sub>2</sub>O. The rest of the volumes in the PCR mix corresponded to the standard PCR mix on the [[https://2011.igem.org/Team:NTNU_Trondheim/Protocols protocols page]]. |
| + | Concentration was measured after the PCR: | ||
| + | {|border="1" | ||
| + | !Sample | ||
| + | !Concentration [ng/µl] | ||
| + | |- | ||
| + | |0.5 µl template DNA | ||
| + | |118.8 | ||
| + | |- | ||
| + | |5 µl template DNA | ||
| + | |103.4 | ||
| + | |} | ||
| + | To test if we had the right product, 5 µl of both samples was investigated using gel electrophoresis. | ||
| + | The PCR-amplified RelA was digested with EcoRI and SpeI and purified. Concentration of the sample isolated from 0,5 µl template was 5,9 ng/µl while the one isolated from 5 µl was 4,3 ng/µl. The RelA insert cut with EcoRI and SpeI was ligated to Term backbone (cut with EcoRI and XbaI). | ||
{{:Team:NTNU_Trondheim/NTNU_footer}} | {{:Team:NTNU_Trondheim/NTNU_footer}} | ||
Revision as of 21:06, 27 July 2011

Lab Journal
|
|
|
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Thursday 23/6
Lab equipment was prepared: Pipette tips, 1.5 ml tubes, toothpicks, SOC, LA with 100 µg/ml ampicillin/ 100 µg/ml spectinomycin (from 1988). Recipies are given in recipies section.
Friday 24/6
Biobricks were taken out from kit by resuspending them in sterile water followed by transformation to E.coli DH5 alpha. The biobricks that were taken out are given below:
| Abbreviation | Name | Part number | Resistance |
|---|---|---|---|
| P1 | rrnB P1 | BBa_K112118 | Spec |
| lambda pR | lambda pR mod | BBa_R0051 | Amp |
| RFP | TetR + p(TetR)+RFP | BBa_K092600 | Amp |
| Lux | TetR + p(TetR)+RFP+luxI | BBa_K092700 | Amp |
Saturday 25/6
Plates with transformants were moved to fridge.
Sunday 26/6
Transformants were inoculated in 3 ml LB + proper antibiotics to prepare for isolation of plasmid.
Monday 27/6
Plasmids that contained P1, lambda pR, RFP and Lux biobricks were isolated by using miniprep kit from Promega.
Consentrations of the biobricks were measured and were as follows:
| Biobrick | Concentration (ng/µl) |
|---|---|
| P1 | 98.4 |
| lamda pR | 43.8 |
| RFP | 141.8 |
| Lux | 62.8 |
Transformants from Lux/RFP constructs were not red as expected. It was suggested that the phenotype could be explained by presence of TetR in the system. Since tetracycline (Tc) is an inhibitor of TetR we tried to grow the transformants in sublethal consentrations of Tc. E. coli with RFP were grown in 3 ml LB with 0, 0.1, 0.3, 0.6, 1.0, 1.5 and 100 µg/ml Tc.
Tuesday 28/6
It's Jon's birthday! Happy day!
Growing the bacteria in Tc did not result in red color of the culture
lambda pR was ligated to RFP backbone:
- RFP and Luc construct were cut with EcoRI and XbaI.
- lambda pR was cut with EcoRI og SpeI
Restriction digestion was done using [http://partsregistry.org/Help:Protocols/Restriction_Digest iGEM's protocol]
Since the restriction digestion gave unexpected fragments and we had not yet seen any pigmentation, we started looking for other designs that could give the result that we wanted. We came up with a different design that used pLac promoter and lacI repressor instead of the pTet/TetR system.
To start constructing the alternative system 3 new biobricks were transformed to E. coli:
| Abbreviation | Name | Part number |
|---|---|---|
| LacI | LacI + RBS | BBa_J24679 |
| pLac | Lac promoter hybrid | BBa_R0011 |
| mCherry | mCherry + RBS + term | BBa_J06702 |
Wednesday 29/6
Lambda Pr, RFP and Lux was cut:
- RFP with XbaI and PstI
- Lux with XbaI and PstI
- Lambda Pr with SpeI and PstI
The cutting gives RFP and Lux as insert and Lambda pR the backbone.
The restriction fragments were separated on agarose gel:
The restriction fragments for lambda pR were as expected but not for RFP or Lux.
Lambda pR was isolated from gel (2110 nt) by using DNA gel extraction kit.
DNA concentration of lambda pR was measured to 5.2 ng/µl
Biobricks that were transformed yesterday were inoculated in 3 ml LB with proper antibiotics to prepare for isolation of plasmids.
Anders ordered a biobrick that should contain LacI + RBS + double terminator from iGEM HQ: BBa_K292006
Thursday 30/6
Plasmid was isolated from lac-design biobricks. Concentration of DNA:
| Biobrick | Concentration (ng/µl) |
|---|---|
| pLac | 16.5 |
| lacI | 37.1 |
| mCherry | 36.7 |
pLac biobrick was cut with SpeI and PstI mCherry was cut with XbaI and PstI
Makes pLac backbone and mCherry insert
Tried direct ligation of restriction fragments using following reaction mix:
| Compound | Amount (µl) |
|---|---|
| H20 | 11 |
| Backbone | 2 |
| Insert | 2 |
| T4 ligase buffer | 2 |
| Ligase | 1 |
Reaction mix were mixed and incubated 30 min at 16C. Heat destruction of enzymes for 20 min at 80C. 2 µl of ligation mix were used for transformation. Red colonies were to be selected from transformation. Mix, spin down, 30 min 16C, heat kill 20 min 80C. Transform 2µL. Select for red colonies.
The rest of the ligation mix were separated on agarose gel and mCherry insert (895 bp) and pLac backbone (2116 bp) were isolated from gel using Quiaquick Gel Extraction Kit. DNA concentration was measured:
| Biobrick | Concentration (ng/µl) |
|---|---|
| pLac | 8.5 |
| mCherry | 2.1 |
The directly ligated restriction fragments transformed to E. coli were plated out on LA + Amp and LA + IPTG and grown ON.
The biobrick that has been ordered (BBa_K292006) was investigated and was found to be identical to the biobrick that we were planning to make, given that the sequence is as given in the registry.
relA
The nucleotide sequence of relA (codes for protein that synthesize ppGpp) was found in database. One hickup in making this gene a biobrick; there is a PstI site inside the gene (at 1383-1388 nt if added prefix). To overcome this problem we could use partial digestion or we could introduce a silent mutation that removes the restriction site. By changing CTGCAG → CTACAG or CTGCAA this could be done.
LacI-negative E.coli
To make the lac-system work we should use a strain that is lacI-negative. According to [http://partsregistry.org/Part:BBa_K177038 another iGEM group] the strain Top10 is lacI-negative.
Friday 1/7
mCherry insert and pLac backbone was isolated from gel using Quiquick Gel Extraction kit and then ligated using the following reaction mix:
| Compound | Amount (µl) |
|---|---|
| Insert | 14 |
| Backbone | 3 |
| T4 ligase buffer | 2 |
| Ligase | 1 |
The ligation mix was ligated at 16C for 1 hour.
Ligation mix was then transformed to LA + Amp, LA + Amp + IPTG and LA + Amp + more IPTG
Two new biobricks were also transformed and plated out on LA + Amp:
| Abbreviation | Biobrick | Part name |
|---|---|---|
| GFP | GFP + LVA | BBa_K082003 |
| RBS | Ribosome binding site | BBa_B0034 |
Monday 4/7
Plasmid isolation of GFP, RBS and LacP+mCherry. Concentrations were measured.
| Biobrick | Concentration (ng/µl) |
|---|---|
| GFP | 60.1 |
| RBS | 16.2 |
| pLac + mCherry | 29.1 |
Restriction cutting of RBS and GFP:
- RBS: SpeI and PstI making it the backbone
- GFP: XbaI and PstI making it the insert.
Gel Extraction:
Extracted RBS backbone, and GFP insert form the gel.
| Biobrick | Concentration (ng/µl) |
|---|---|
| GFP | 3.0 |
| RBS | 4.6 |
Plating:
Plating of pLac + mCherry on amp plates, amp plates with additional IPTG (7µL), amp + IPTG plates and amp + IPTG plates with additional IPTG (7 µL). Incubating on 30 and 37 degrees.
Tuesday 5/7
Ligation of RBS backbone and GFP insert.
- 10μL Ligation Mix
- 1.0 μL 10X T4 ligase buffer
- 6:1 molar ratio of insert to vector (~10ng vector)
- Add (8.5 - vector and insert volume)μl ddH2O
- 0.5 μL T4 Ligase
Used 2,5 µL RBS backbone and 6 µL GFP insert. Incubating at 16 degrees celsius, then heating at 80 degrees celsius.
PCR amplification of rrnB P1 from BioBrick
Resuspend primers in dH20. Primers were designed by the team, and synthesized by [http://www.sigmaaldrich.com Sigma-Aldrich]. Primers will amplify the whole rrnB P1 BioBrick (rrnB.FWD and REV), and only the assumed promoter sequence (pro.FWD and REV), and will add normal prefix and suffix instead of BBb format.
Primers used:
| Primer | Type | Sequence |
|---|---|---|
| rrnB P1 F | Forward | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGACGTATCCTACGCCCGTGGT |
| rrnB P1 R | Reverse | GTTTCTTCCTGCAGCGGCCGCTACTAGTACGCCTTCCCGCTACAGAGTCA |
| proL F | Forward | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGCCTCTTGTCAGGCCGGAATAACTCC |
| proL R | Reverse | GTTTCTTCCTGCAGCGGCCGCTACTAGTAGCGGCGTGTTTGCCGTTGTT |
PCR mix:
Work on ice
- Dillute P1 DNA 1µL + 9µL dH2O → ( 9,84 g/µL P1 DNA)
- 2 x PCR tubes 0,5 µL DNA
- 5 µL 10x PCR buffer 2 (w/MgCl)
- 0,5 µL 10 mM dNTPs
- 1 µL fwd primer (rrnBL og proL)
- 1 µL rev primer (rrnBL og proL)
- 0,5 µL polymerase
- 41,5 µL H20
Heat cycles:
Heated lid 103C
- Initial denaturation: 94C 2 min
- Denaturing: 94C 30 s
- Annealing 58C 30 s
- Elongation 72C 60 s
- Repeat 2-4 34 times
- Final elongation 72C 7 min
- Cooling 4 C HOLD
Running products on 1,5 % agarose to find short fragment (promoter sequence). Should give 552 bp (rrnB) and 129 bp (proL).
The products were not so good, so we are running a new PCR tomorrow with different scheme.
Inoculated "LacI with RBS", TERM and LacP+MC biobrick. LacP+MC with and without IPTG and incubating at 30 and 37 degrees celsius.
Wednesday 6/7
Running PCR with rrnBS and L and proS and L on the rrnB P1 BioBrick. 5 cycles of 58C annealing temp, and 20 cycles of 68C annealing temp to try adjust for the massively changed annealing temperatures when primers with pre- and suffixes bind to PCR products. The PCR products where separated by gel electrophoresis and cut out, ready for gel extraction tomorrow.
Isolated plasmids containing the "LacI with RBS" and "TERM" biobricks.
| Biobrick | Concentration (ng/µl) |
|---|---|
| LacI with RBS | 26.0 |
| TERM | 29.9 |
Restriction cutting of "LacI with RBS" with EcoRI and SpeI, and "TERM" with EcoRI and XbaI. The fragments where separated using gel electrophoresis.
The "LacI with RBS" insert and the backbone containing "TERM" where cut out of the gel, and extracted from it.
| Biobrick | Concentration (ng/µl) |
|---|---|
| LacI with RBS insert | 1.6 |
| TERM backbone | 3.9 |
RBS+GFP transformants where inoculated in 3 mL LB+amp.
Thursday 7/7
Ligation of TERM backbone and LacI with RBS insert extracted from gel 6/7.
10μL Ligation Mix:
- 1.0 μL 10X T4 ligase buffer
- 6:1 molar ratio of insert to vector (~10ng vector)
- Add (8.5 - vector and insert volume)μl ddH2O
- 0.5 μL T4 Ligase
Used 2,5 µL TERM backbone and 6 µL LacI with RBS insert. Incubated at room temperature for 30 minutes, then heated at 65 C for 15 minutes.
- Plasmid with GFP+RBS was isolated. Concentration was 26.7 ng/µl
Restriction cutting:
rrnB P1 BioBrick
The rrnB P1 BioBrick found in the distribution was in a different format (BBb), and was therefore not compatible with the other BioBricks we are using. PCR was used to amplify the BioBrick itself, adding the prefix and suffix found [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication here] making it compatible with the other parts, but losing the first 10 bp. We also amplified only the assumed promoter sequence, with the pre/suffixes.
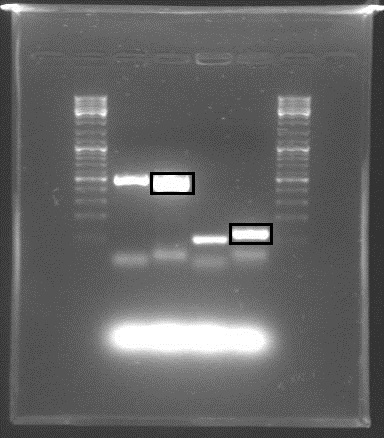
Gel electrophoresis with the PCR products showed that DNA of the right sequence length had been amplified, as shown in the figure. The full-length rrnB P1 amplicon had an approximate size of 500 bp, and the shorter promoter-only amplicon was approx. 100 bp. The shorter fragments shown were created using shorter primers with only X and S restriction sites, in case the long primers failed.
The amplicons with the complete pre/suffixes were cut out, as shown in the figure, and purified with QIAquick Gel Extraction Kit.
Both promoter variants were cut with E and P and also with E and S to create insertable pieces. Our RBS+GFP BioBrick was cut with E and X to create a backbone, and the linearized backbones psB1A3 and psb1C3 were cut to accomodate the cut PCR products.
Friday 8/7
It's Friday, we excited...
- The following ligations were made:
| Testing activity | Ligate in BB plasmid |
|---|---|
| rrnBL+mCherry | rrnBL+backbone pSB1A3 |
| rrnBL+(RBS+GFP) | rrnBL+backbone pSB1C3 |
| ProL+mCherry | ProL+backbone psB1A3 |
| ProL+(RBS+GFP) | ProL+backbone psB1C3 |
- RBS + GFP was cut with Xbal and PstI, and the restriction fragments were separated on agarose gel. The insert fragment RBS + GFP were as expected (ca 750 bp), and the fragment was cut out from gel and extracted by using DNA gel extraction kit. The DNA concentration of RBS + GFP insert was measured to 2.1 ng/µl.
PCR amplification of relA from chromosomal DNA
Resuspend primers in dH20. Primers were designed by the group and synthesized by Sigma-Aldrich. Primers will amplify the whole relA gene from E. coli and add normal prefix and suffix (biobrick standard).
Primers used:
| Primer | Type | Sequence |
|---|---|---|
| relA F | Forward | GTTTCTTCGAATTCGCGGCCGCTTCTAGAGATGGTTGCGGTAAGAAGTGCACA |
| relA R | Reverse | GTTTCTTCCTGCAGCGGCCGCTACTAGTACTAACTCCCGTGCAACCGACG |
PCR mix:
Same as used earlier.
Heat cycles:
Heated lid 103C
- Initial denaturation: 94C 2 min
- Denaturing: 94C 30 s
- Annealing: 58C 30 s
- Elongation: 72C 150 s
- Repeat 2-4 4 times
- Denaturing: 94C 30 s
- Annealing: 68C 30s
- Elongation: 72C 150 s
- Repeat 6-8 19 times
- Final elongation 72C 7 min
- Cooling 4 C HOLD
PCR products where put in the freezer for further work tomorrow.
Saturday 9/7
Transformed new promoter system for our relA gene from biobrick plate and plated out on Kam plates:
| Abbreviation | Biobrick | Part name |
|---|---|---|
| pBAD | Inducible pBad/araC promoter | BBa_I0500 |
Changed medium for our TOP10 culture
Separated PCR products of the relA gene on gel and used QIAquick Gel Extraction Kit for cleanup. Concentration: 3 ng/µL
Monday 11/7
The new relA biobrick was digested with EcoRI and PstI for insert in plasmid backbone pSB1A3. The following reaction mix was used:
- 17 μl DNA
- 2 μl buffer 3
- 1 μl EcoRI
- 1 μl PstI
- 0.5 μl BSA
- Incubation on a 37 °C water bath for 30 min
- Heat kill (80 °C for 20 min)
Then the digested relA was ligeted with linearized pSB1A3 using the iGEM procedure for ligeting linearized plasmid backbones. The procedure can be found [http://partsregistry.org/Help:Protocols/Linearized_Plasmid_Backbones here].
pSB1A3 containing relA was then transformed to competent DH5 α cells and incubated overnight.
Plasmid Isolation:
| Biobrick | Concentration (ng/µl) |
|---|---|
| rrnBL in pSB1C3 | 18,9 |
| ProL in pSB1C3 | 41,6 |
| rrnBL in pSB1A3 | 22,8 |
| ProL in pSB1A3 | 29,6 |
| rrnBL+GFP | 47,5 |
| proL+GFP | 40,8 |
| rrnBL+MC | 85,3 |
| ProL+MC | 48,1 |
| RBS+LacI+Term | 53,9 |
| LacP+GFP | 26,6 |
| Lambda Pr | 38,4 |
Restriction Digestion:
Digested rrnBL in pSB1A3 with SpeI and PstI and the LacI construct (RBS+LacI-TERM) with Xba1 and PstI
The resulting fragments were separated using gel electrophoresis. rrnBL backbone and LacI insert were cut out of the gel and stored at -20
Top10 cells: We would like to use top10 cells because it lacks a wild type LacI system. The top10 cells we have already contains a plasmid with Kan resistance. We have been incubating the top10 cells for a couple of days without selection so that some of the cells will loose the plasmid. Today we plated several dillutions of culture in order to get single colonies. The dilutions where 1:10, 1:100, 1:1000, 1:10000 and 1:100000. The plates were incubated over night.
Tuesday 12/7
Colonies from TOP10 plates were transferred to new plates with LA and LA + Kan to check wether the cells had lost their plasmid or not.
Since the first GFP biobrick we tried did't work properly, we transformed two new variants of GFP. In addition, we also transformed the biobrick coding β-galactosidase, to be able to measure activities of different promotors by setting the promotor in front of the β-galactosidase biobrick, induce the promotor, destroy the cell and look at the absorbance when we add ONPG.
| Abbreviation | Name | Part number | Resistance |
|---|---|---|---|
| GFP 2 | GFP without degradation tag | BBa_E0040 | Amp |
| GFP ssra | GFP with ssra degradation tag | BBa_E0044 | Amp |
| LacZ | β-galactosidase | BBa_I732019 | Amp |
The fragments cut out of the gel on monday were extracted. The resulting consentrations were 1 ng/µL of rrnBL backbone and 1,6ng/µL of LacI insert. The parts were then ligated using 3,5µL backbone and 5µL insert. Then transformation and incubation on plate followed.
Wednesday 13/7
To see if our rrnB P1 + mCherry construct is working properly, it was made a dilution series from the cell culture, and the concentrations 10-3, 10-4, 10-5 and 10-6 was plated out on LA+Amp plates.
The plasmids containing the pBad/AraC promotor, the relA gene and the rrnB P1 + mCherry construct was isolated using the miniprep kit from Promega. The concentrations of the plasmids was measured.
| Biobrick | Concentration [ng/μl] |
|---|---|
| pBad | 24,6 |
| RelA | 80,8 |
| P1+mC | 45,8 |
The P1+Lac construct, the LacZ gene and the two new GFP biobricks were inoculated.
RelA-plasmid was digested with E and S to yield RelA insert (2234 bp)and was separated from its backbone (2155) and the larger piece was cut out with surgical precision.
RelA DNA was purified from the gel-slab and ligated into TERM backbone. The ligation mix was transformed into DH5-alpha cells.
Thursday 14/7
Plasmids; pBAD/AraC, P1+Lac construct, the LacZ gene and the two new GFP biobricks were isolated, giving the following concentrations:
| BioBrick | Concentration ng/µL |
| pBAD/AraC | 24,6 |
| lacZ | 76,1 |
| GFP2 | 32,4 |
| GFP ssra | 59,8 |
| P1+LacI | 17,3 |
The lacZ construct was digested with E+X to yield a backbone for rrnB insert digested with E+S. The backbone digest was isolated from gel and saved.
GFP2 and GFP ssra were digested with X+P, P1+lacI was digested with E+S, lacP+mCherry with E+X, and RBS+lacI+TERM with E+S. The fragments were separated and isolated from electrophoresis, and the DNA was purified from the gel-pieces.
The concentrations of DNA after gel purification were as follows:
Friday 15/7
The LacZ backbone gel cut-out was isolated using QIAquick Gel Extraction Kit.
Then, the following ligation mixes was made using the alternative ligation procedure on the protocols page.
| Insert | Backbone | Insert added [µl] | Backbone added [µl] | dH2O added [µl] |
|---|---|---|---|---|
| P1+LacI | LacP+mCherry | 1 | 7.5 | 0 |
| RBS+LacI+TERM | LacP+mCherry | 4.5 | 4 | 0 |
| GFP2 | RBS | 6 | 2.5 | 0 |
| GFP ssra | RBS | 6.5 | 2 | 0 |
| rrnB P1 | RBS+LacZ+TERM | 1 | 3.6 | 3.9 |
2 µl of each ligation mix was transformed to competent DH5α cells. The cells were plated out on LA + Amp plated and incubated on 37°C.
Saturday 16/7
Transformed pBAD2+relA and religation to competent cells.
Plated out TOP10 cells in different dilutions (1:10 - 1:1000000).
Sunday 17/7
Colonies from TOP10 plates were transferred to new plates with LA and LA + Kan to check wether the cells had lost their plasmid or not.
Inoculated the following constructs; pBAD2+relA; P1+LacI+LacP+mCherry; RBS+LacI+TERM+LacP+mCherry; RBS+GFP2; RBS+GFPssra; rrnB P1+RBS+LacZ+TERM.
Monday 18/7
The following plasmids were isolated:
| Biobrick | Concentration (ng/µl) |
|---|---|
| RBS+GFP2 | 59,0 |
| RBS+GFP ssrA | 61,6 |
| pBad+RelA | 55,0 |
| rrnB P1+RBS+LacZ+Term | 35,5 |
| RBS+LacI+TERM+LacP+MC | 38 |
| rrnB+RBS+LacI+TERM+LacP+MC | 22,6 |
RBS+GFP2 and RBS+GFP ssrA were cut with Xba1 and PstI. the resulting fragments were separated on a gel and the insert fragments were cut out and extracted.
| Insert | consentration |
|---|---|
| GFP2 | 3,1 |
| GFP ssrA | 2,8 |
The GFP's were ligated with promotors as follows:
| Insert | µL | Backbone | µL |
|---|---|---|---|
| GFP2 | 7,3 | LacP | 1,2 |
| GFP2 | 6,5 | Lambda Pr | 2,0 |
| GFP2 | 3,5 | rrnB (P1) | 5 |
| GFP ssrA | 7,5 | LacP | 1,0 |
| GFP ssrA | 6,7 | Lambda Pr | 1,8 |
| GFP ssrA | 3,5 | rrnB (P1) | 5 |
Religations of the backbones were also made (Water instead of insert).
Different ways to stress the cells were discussed, and we decided that we could try making them stressed both by growing them in a nullmedium, a minimal a minimal medium, and in normal medium, but on higher and lower temperatures than 37°C Nullmedium and minimal medium were prepared and autoclaved.
Nullmedium:
- 0.9 % NaCl (9 g)
- 1000 ml dH2O
M9 minimal medium:
- 0.4 % glucose (4 g/l)
- 2 mM MgSO4*7H2O (0.4922 g/l)
- 0.1 mM CaCl2 (0.0111 g/l)
- 8.49 g/l Na2HPO4*2H2O
- 3.84 g/l K2HPO4
- 0.5 g/l NaCl
- 1 g/l NH4Cl
- 1000 ml dH2O
- pH was adjusted to 7.4 using HCl
In addition, 2 L LA medium was prepared using the recipe on the protocols page, to make LA plates with kanamycin, ampicillin, and without antibiotics.
The RBS+LacI+TERM+LacP+mCherry inoculum was clearly red, indicating expression of mCherry and showing that it could be used as an indicator in liquid culture as well.
P1+LacI+LacP+mCherry was cut with three different enzyme pairs to test for the presence of the proper BioBricks, and size of the complete construct. The digestions are shown below.
| Enzyme-pair | Expected fragments |
|---|---|
| EcoRI, SpeI | 2056bp, 2771bp |
| BsaAI, HpaI | 4083bp, 744bp |
| BstBI, SpeI | 3284bp,934bp |
Hopefully, the digestions will indicate a construct as shown in the plasmid map.
Tuesday 19/7
Prepared new M9 media, as the one yesterday was a complete FAIL. The media was prepared as shown in the protocols.

rrnB+LacZ and pBad+RelA was testcutted with different combinations of restriction enzymes:
- rrnB+LacZ:
- Cutted with HindIII and SacI → 4305 bp + 2615 bp
- Cutted with EcoRI and SpeI → 3754 bp (insert) + 3166 bp (backbone)
- Cutted with NdeI and SacI → 1017 bp + 5903 bp
- pBad+RelA:
- Cutted with AflII and SexAI → 3201 bp + 2407 bp
- Cutted with EcoRI and XbaI → 5652 bp (backbone)
We then did gel electrophoresis for the five cut samples for rrnB+LacZ and pBad+RelA and the three samples of rrnB+LacI+TERM+LacP+mCherry that was cut yesterday with three different combination of enzymes. For the first gel, containing the three samples of rrnB+LacZ and the sample of pBad+RelA cut with AflII and SexAI, it was not possible to see any distinct difference between the sizes of the DNA pieces, but for the second gel, containing pBad+RelA cut with EcoRI and XbaI and the three samples of rrnB+LacI+TERM+LacP+mCherry, the gel looked better. pBad+RelA cut with EcoRI and XbaI gave a backbone we intend to use later, so this was cut out of the gel.
Wednesday 20/7
rrnB+LacZ and pBad+RelA was test-cut again with the same combination of enzymes that we did yesterday.
We also transformed cells with the rrnB+lacZ construct that was test-cut, in case we need more DNA.
We used gel electrophoresis to separate the test-cut plasmids. The pBad+relA-sample cut with EcoRI and XbaI gave a backbone, and was cut out of the gel, and the rrnB+lacZ-sample cut with EcoRI and SpeI gave an insert, and was also cutt out of the gel. The rrnB+lacZ-sample cutt with EcoRI and SpeI contained both an insert (3754 bp) and a backbone we are not going to use (3166 bp). Since these two DNA fragments were very similar in size, the gel electrophoresis had to proceed for about four hours.
The six parallell RBS+lacI+TERM_lacP+mCherry plasmid and P1+lacI+TERM+lacP+mCherry were test-cut as follows.
| Enzymes | Fragments |
|---|---|
| EcoRI, NcoI | 2475 + 1851 |
| BsaAI, HpaI | 312 + 3894 |
| EcoRI, SpeI | 2056 + 2771 |
| BsaAI, HpaI | 4083 + 744 |
| BstBI, SpeI | 3284 + 934 |
lacP+GFP2 and lacP+GFPssra transformants were all glowing under UV-light. rrnBP1+GFP2 and rrnBP1+GFPssra were also glowing, but less bright than the lacP constructs. lambdaP constructs were not visibly glowing.
Colonies from the rrnb P1 and lambdaP GFP-constructs were inoculated for plasmid isolation.
Thursday 21/7
Stress-testing the final construct
Pre-cultures for biological stress-tests were inoculated in LB+amp at 37 C at 10:35. At 14:15 the cultures were pelleted down at 2000 rpm for 5 min and resuspended in remaining liquid.
One of the stress-experiments is to induce Heat-Shock. One culture will e grown at 42 C, and one will be grown at 37 C but will be heat-shocked at 42 C several times. The Heat-Shock cultures were inoculated at 14:35. Heat-shock was induced at 14:55 until 15:25.
Two starvation stress-experiments were also performed, where the cells were resuspended and inoculated in M9 minimal media and in 0,9% NaCl. The incubated constructs included the P1-lacI+lacP-mCherry complete construct, and also the construct lacking the rrnB P1 promoter. Controls were also incubated in regular LB at 37C. The cultures were left over night in shaking incubators.
GFP-construct plamids were isolated;
| Construct | Concentration (ng/µL) |
|---|---|
| rrnB+GFP2 (1) | 23,3 |
| rrnB+GFP2 (2) | 24,1 |
| rrnB+GFPssra (1) | 19,7 |
| rrnB+GFPssra (2) | 23,2 |
| lamdbaP+GFP2 | 27,0 |
| lamdaP+GFPssra | 31,4 |
The isolated plasmids were digested with E and S to yield inserts, to eventually put in front of our construct to be able to have green bacteria (in UV) as a positive indicator. In addition the plasmids were digested with NcoI and StuI, to test for GFP insert (NcoI) and the ssra-tag (StuI). The fragments should be as follows:
| Construct | Enzyme pair | fragments |
|---|---|---|
| rrnB P1+GFP2 | EcoRI, SpeI | 1242(insert) + 2132 |
| NcoI, StuI | 3374 | |
| rrnB P1+GFPssra | EcoRI, SpeI | 1281(insert) + 2132 |
| NcoI, StuI | 551 + 2682 | |
| lambda Pr+GFP2 | EcoRI, SpeI | 798(insert) + 2056 |
| NcoI, StuI | 2854 | |
| lambda Pr+GFPssra | EcoRI, SpeI | 837(insert) + 2056 |
| NcoI, StuI | 2342 + 551 |
rrnB P1 characterization construct:
The gel pieces that were cut out yesterday, containing rrnB+LacZ insert and pBad+RelA backbone, were purified using the QIAquick gel extraction kit, and the concentrations were determined:
| Biobrick | Concentration [ng/µl] |
|---|---|
| pBad+RelA | 6.7 |
| rrnB+LacZ | 4.1 |
The pBad+RelA backbone and the rrnB+LacZ insert were ligated using the alternative ligation procedure on the protocols page. A religation mixture was also made. The following volumes were used:
| Backbone | Insert | Volume backbone [µl] | Volume insert [µl] | Volume water [µl] |
|---|---|---|---|---|
| pBad+RelA | rrnB+LacZ | 1.49 | 7.01 | 0 |
| pBad+RelA | - | 1.49 | - | 7.01 |
The ligated plasmid and a religation was transformed to competent E.coli DH5α cells, and incubated on 37 °C.
Friday 22/7
| Construct | Enzyme pair | Applied to well | Fragment sizes (bp) |
|---|---|---|---|
| rrnB P1+GFP2 1 | EcoRI, SpeI | 3 | 1242(insert) + 2132 |
| NcoI, StuI | 2 | 3374 | |
| rrnB P1+GFP2 2 | EcoRI, SpeI | 5 | 1242(insert) + 2132 |
| NcoI, StuI | 4 | 3374 | |
| rrnB P1+GFPssra 1 | EcoRI, SpeI | 7 | 1281(insert) + 2132 |
| NcoI, StuI | 6 | 551 + 2682 | |
| rrnB P1+GFPssra 2 | EcoRI, SpeI | 11 | 1281(insert) + 2132 |
| NcoI, StuI | 10 | 551 + 2682 | |
| lambda Pr+GFP2 | EcoRI, SpeI | 12 | 798(insert) + 2056 |
| NcoI, StuI | 13 | 2854 | |
| lambda Pr+GFPssra | EcoRI, SpeI | 14 | 837(insert) + 2056 |
| NcoI, StuI | 15 | 2342 + 551 |
Top10 cells were plated out on agar plates were a grid was made. They were plated out on both LA plates and LA+Kan plates with equally looking grids, and will be investigated later to find out if any of the cells have lost their plasmid.
Two samples of P1+GFP2, two samples of P1+GFPssra, one sample of λPr+GFP2 and one sample of λPr+GFPssra, all cut with E+S yielding inserts was run on gel, and DNA from the gel pieces was isolated using the QIAquick gel extraction kit.
Concentration was measured:
| Biobrick | Concentration [ng/µl] |
|---|---|
| P1+GFP2 1 | 2.3 |
| P1+GFP2 2 | 1.9 |
| P1+GFPssra 1 | 3.3 |
| P1+GFPssra 2 | 2.7 |
| λPr+GFP2 | 3.0 |
| λPr+GFPssra | 4.3 |
Saturday 23/7
We have finally made E. coli TOP10 lose its plasmid!!
Sunday 24/7
The transformations looked good, with many colonies on each plate. Negative control and religation control had 0 colonies. 4 colonies from each transformation were inoculated in 3 ml LB + Amp to prepare for plasmid isolation.
Monday 25/7
Colonies of TOP10 cells who had lost its plasmid (from saturday 23/7) were transferred to new plates with LA and LA + Kan, as well as LB medium.
We found out that our RelA constructs were missing RBS :( We need to take some steps back and introduce an RBS to RelA+Term. This was done by cutting RelA+Term with X+P making it insert. Sinve RelA has a PstI site inside the gene, the plasmid needs to be partially digested with PstI. It was digested with X over night to prepare for partial digestion tomorrow.
Tuesday 26/7
1 ml TOP10 cells in LB medium grown over night were mixed with glycerol (400 µl, 60%) and stored in the freezer for use later.
1L M9 minimal medium were made.
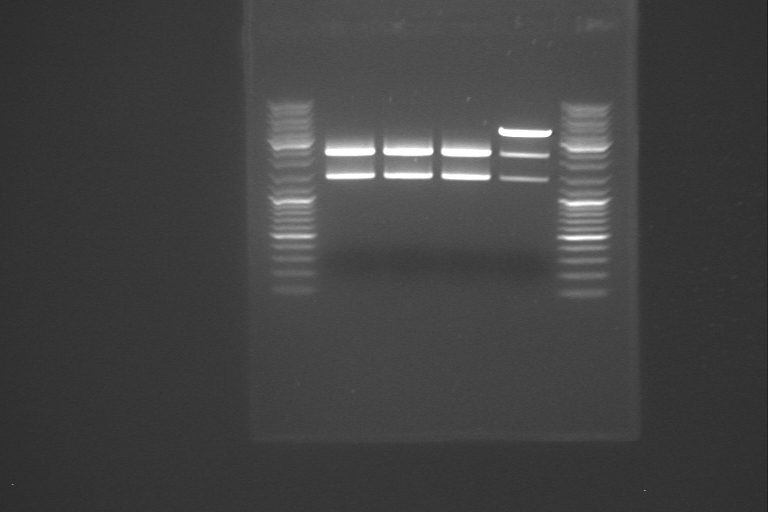
The samples from yesterday was partially digested with PstI using a dilution series. The first sample was made according to the procedure for digestion cutting on the protocols page. The next two samples was also made according to this procedure, but with 10 µl less dH2O than usual. After adding PstI to the first sample, it was mixed, and 10 µl was transferred to the second sample. The second sample was mixed, and 10 µl transferred to the third sample. The sample that was digested with XbaI and PstI was separated on gel alongside with the three samples from the dilution series from the partial digestion. The gel picture didn't make sense, so we started concidering different reasons why the fragments were not as expected. We looked in the lab journal, to find out what had been done about RelA earlier, and we found out that when our PCR product was inserted in a plasmid, PstI and EcoRI was used, without partial digestion. It is likely that RelA was cut in two pieces when it was inserted in the plasmid, and that this is the reason why the fragments on the gel was smaller than expected. If this assumption is correct, we have to start almost all over again, since it is most likely that the relA gene was destoyed in the first cutting step, where it was cloned into the plasmid.
Wednesday 27/7
RelA was amplified using PCR. The PCR product of our first amplification of RelA served as template DNA. Since the concentration of the template DNA was low, we amplified two different samples of it. In the first sample, we added 0.5 µl template DNA and 41.5 µl dH2O, and in the second sample, we used 5 µl of template DNA and 37 µl of dH2O. The rest of the volumes in the PCR mix corresponded to the standard PCR mix on the [protocols page]. Concentration was measured after the PCR:
| Sample | Concentration [ng/µl] |
|---|---|
| 0.5 µl template DNA | 118.8 |
| 5 µl template DNA | 103.4 |
To test if we had the right product, 5 µl of both samples was investigated using gel electrophoresis. The PCR-amplified RelA was digested with EcoRI and SpeI and purified. Concentration of the sample isolated from 0,5 µl template was 5,9 ng/µl while the one isolated from 5 µl was 4,3 ng/µl. The RelA insert cut with EcoRI and SpeI was ligated to Term backbone (cut with EcoRI and XbaI).
 "
"