Commons
PCR
| Name: Sophie
| Date: 25.07.11
|
| Continue from Experiment (Date)
(Name): Commons
|
| Project Name: more linearized backbones (4 different vectors)
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| SB-prep-3P-1
|
| 2.5µl
| Primer dw
| SB-prep-2Ea
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
| PSB 1 A3
PSB 1 C3
PSB 1 K3
PSB 1 T3
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
- 2Min 94°C
- 30s 94°C
- 30s 55°C
- 3min 72°C
- 10min 72°C
step 2,3 and 4 in 35 cycles
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
Labeled PSB 1 A3, PSB 1 C3, PSB 1 K3 and PSB 1 T3 stored in -20°C in last drawer
green light receptor
Testdigest Ligation of CcaS into pSB1K3
Investigators:JULIA
Testdigest
| Continue from Experiment (Date 22.07) Ligation of CcaS into pSB1K3
|
| Project Name: Green light receptor
|
For one reaction you need For Mastermix: 35 samples+2extra
| 4μl
| H2O
| 178μl
| H2O
|
| 1μl
| Buffer, NEB4
| 37μl
| Buffer, NEB4
|
| 1μl
| BSA (10x)
| 37μl
| BSA (10x)
|
| 0,5 μl
| Enzym 1
| 7μl
| EcoRI
|
| 0,5 μl
| Enzym 2
|
|
|
| 3 μl
| DNA
|
|
|
10 μl total volume
Give 3 μl of DNA in an eppi and add 7μl of the mastermix.
Incubate for about 1h at 37°C.
Add 1 μl Loading dye buffer and load the gel.
Picture of 1% gel:
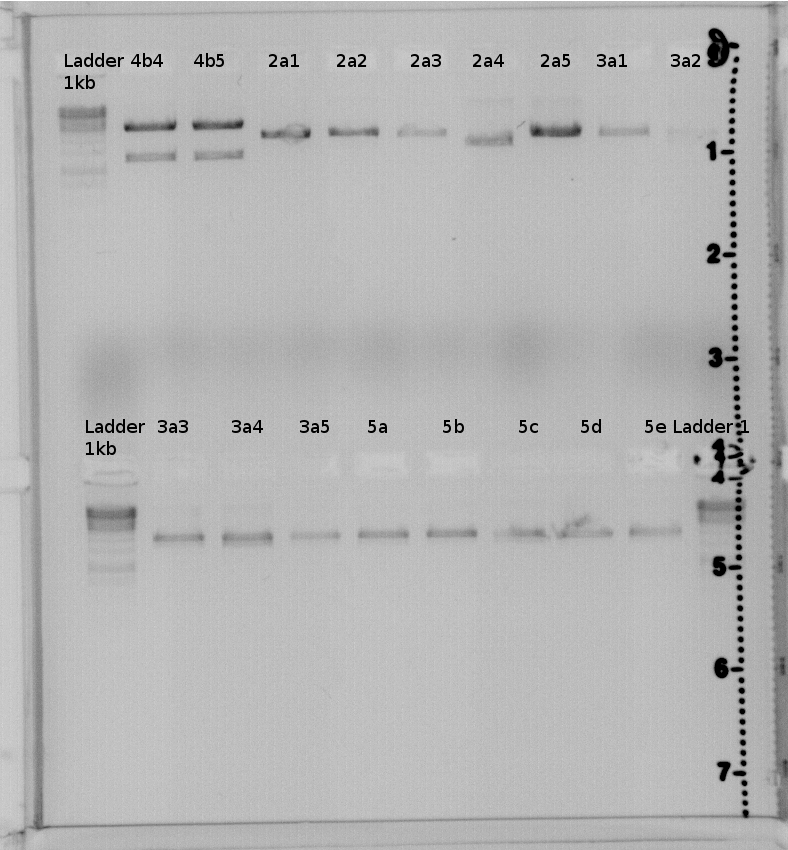

result:4a2,4b2,4b3,4b4,4b5 should have the right insert.
blue light receptor
NAME OF YOUR EXPERIMENT
- Gibson NOT-Gate
Investigators:
- Sophie
Transformation
| Name: Sophie
| Date: 25.07.11
|
| Continue from Date Name
Experiment
|
| Project Name: Blue light (NOT-Gate)
|
Procedure
- take cells from -80°C freezer and put them on ice! (every eppi contains about 400 μl cells)
- thaw cells on ice 20 minutes
- pipette 50 μl cells and 2 μl DNA into eppi still on ice!
- Incubate for 30 minutes on ice
- Heat at 42°C for 60 sec
- Incubate on ice for 5 minutes
- Add 200 μl LB Broth
- Incubate for 2 hours at 37°C (cells with lysis cassette at 30°C!!)
- Plate 50 μl and 200μl on two different LB/Agar plates with appropriate antibiotic resistance
Documentation:
Why are you doing this experiment? Name of the sample? Where are they stored? Name the vector with inserts, antibiotika resistance etc.
| We need a NOT Gate for our Blue light system. In this experiment I transformed E.coli with the Part Bba_Q04400 from the iGEM distribution kit. The vector is pSBK3 with Kanamycin resistance. The strain ist named S 45 and will be plated out. The plates will be stored in the incubator.
|
PCR
| Name: Sophie
| Date: 25.07.11
|
| Continue from Experiment: new experiment. We need NOT-Gate and LovTAP with Gibson-overhangs, so 2 PCRs are made
|
| Project Name: Blue Light
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer:
|
| 2.5µl
| Primer fw
| NOT_G_up
LOV_G_up
|
| 2.5µl
| Primer dw
| NOT_G_dw
LOV_G_dw
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
| S35 (BBa_K322999)
S45 (BBa_Q04400)
|
| 0.5 µl
| Phusion (add in the end)
|
|
What program do you use?
"57°C auf 70°C" (first annealing temperature:55°c, after 10 cycles 65°c)
To confirm the PCR-Product has the correct size, load 2 µl of the sample onto an agarose-gel.
How did you label the PCR-Product, where is it stored and what do you do next?
I labelled it with a heart and a star and stored it in the minipreps 3 box.
I will do Gibson assemblz of the two parts next.
DNA-concentration measured with nanodrop:
| sample
| DNA-concentration (ng/μl)
|
| S35
| 75.3
|
| S45
| 27.2
|
 We will repeat the PCr, because of the bad result of S35. We will do a PCR with higher temperature.
We will repeat the PCr, because of the bad result of S35. We will do a PCR with higher temperature.
red light receptor
NAME OF YOUR EXPERIMENT
Investigators:NAME
Lysis cassette
NAME OF YOUR EXPERIMENT
Investigators:NAME
Precipitator
NAME OF YOUR EXPERIMENT
Investigators: NAME
 "
"
 Contact
Contact 














