Team:Paris Bettencourt/Experiments/Methodologies/Integration
From 2011.igem.org
LauradaSilva (Talk | contribs) |
LauradaSilva (Talk | contribs) |
||
| Line 23: | Line 23: | ||
|} | |} | ||
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+ Bacillus subtilis integration vector with RFP cassette | ||
| + | |- | ||
| + | |[[File:pDCPKO with BB-1.png|900px|center|]] | ||
| + | |||
| + | |- | ||
| + | |} | ||
<html> | <html> | ||
Revision as of 21:35, 28 October 2011

Integration plasmid
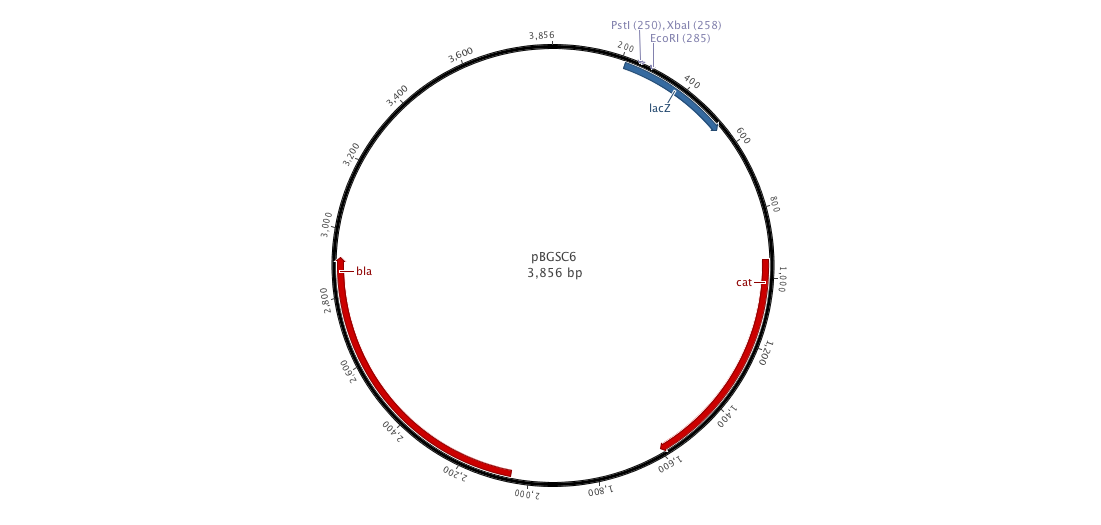
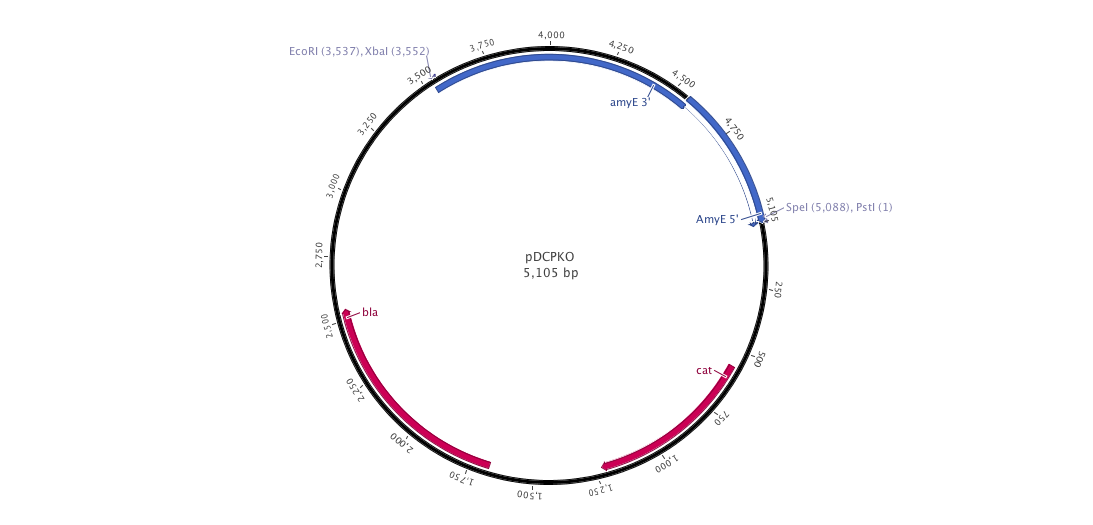
Since we have a lot of problems with the clonning of our parts into replivative plasmid we created, we decided to ;anaged a integrative plasmid which could give us certainly better results since contrary to the replicative plasm we cannot observe a varialibity in its expression. To do that, we created the biobriked plasmid pDCPKO from the Bacillus Genetic Stock Center under the name pBGCS6.
Into this plasmidm we cloned the both AmyE sequences created by the 2008 Imperial College of London team 5 (<partinfo>K143001</partinfo> and <partinfo>K090403</partinfo>)). What we want to do is to clone the whole plasmid into B.subtilis. Thus, we get our next new plasmid pDCPKO which one encodes chloramphenicol acetyl transferase selectable in either E.coli or B.subtilis (chloramphenicol 5μg/ml), β-lactamase selectable in E.coli only (ampicillin 100μg/ml).).
 "
"