Team:Osaka/Tests
From 2011.igem.org
(→Cell viability) |
(→Tests) |
||
| Line 34: | Line 34: | ||
==== Damage detection ==== | ==== Damage detection ==== | ||
* We have assembled a device utilizing GFP as reporter, but did not have time to characterize it properly. The results will be in our final presentation. | * We have assembled a device utilizing GFP as reporter, but did not have time to characterize it properly. The results will be in our final presentation. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
Revision as of 19:25, 28 October 2011
Tests
Cell viability
We performed the UV assay. The cells were plated on respective agar plates at different dilutions, air dried, and then exposed to different doses of UV radiation. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. Please check Protocol for details.
PprM gene could also confer high tolerance to inserted cells.
We expected that all cells inserted each genes could increase its ratio of cell survival, however, two genes, pprI and pprA, couldn't confer tolerance. PprI protein is known as a inducer to genes expression such as recA and pprA. Therefore, expression of only pprI may be ineffective for cell survival. Moreover, inserting heavy gene often causes decline of cell survival.
PprA protein has a function for repairing DNA damaged with blunt end. UV exposure causes thymine dimer, not related to blunt end. We suggest that pprA gene may have no function for repairing DNA damaged by UV but some repairing function for other types of damage such as by chemicals, of cause, radiation.
Fortunately, our result about gene mix(connected each genes) showed high tolerance to UV exposure.
RecA gene could induce high cell viability. RecA protein has key role of SOS response. This result revealed that the cell inserted recA gene can get tolerance against DNA damage.
SOS promoter assay
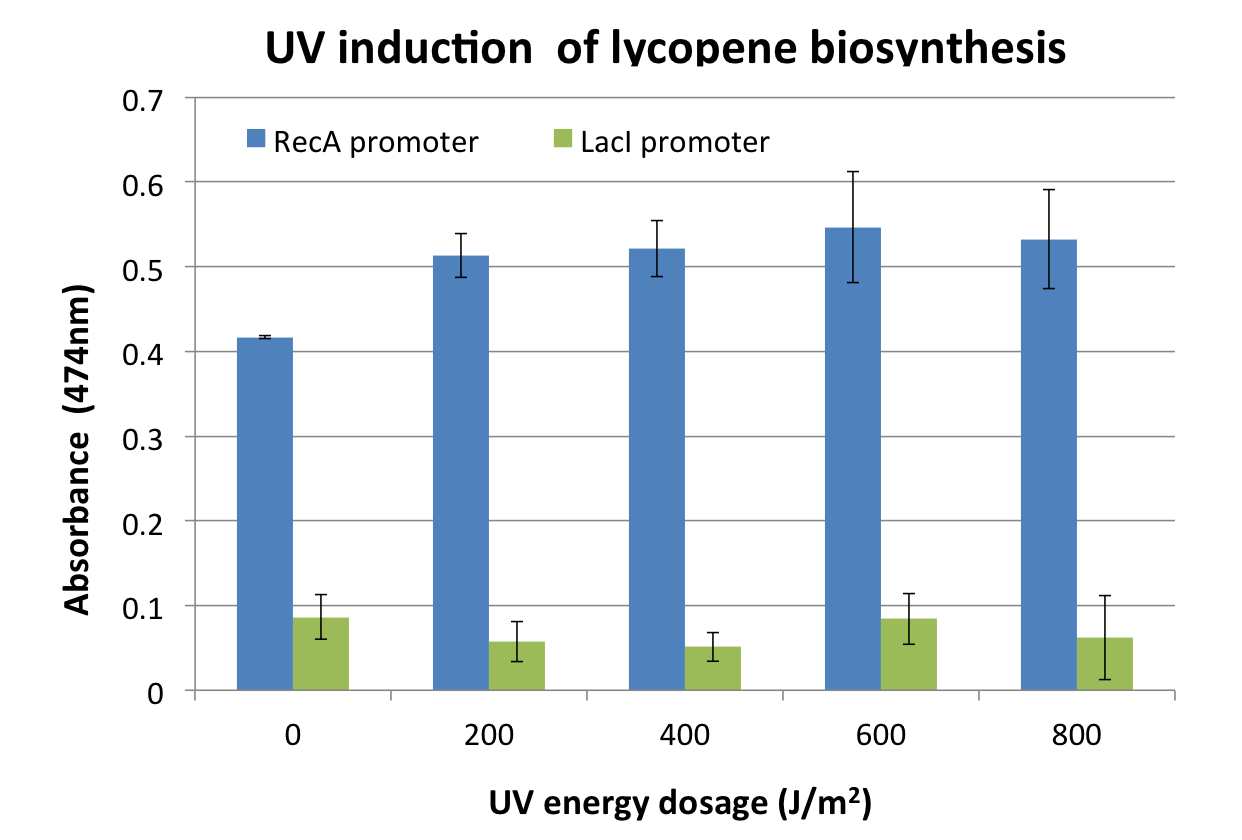
We assayed the promoter of the SOS gene RecA ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]), by attaching a lycopene biosynthesis gene cluster ([http://partsregistry.org/Part:BBa_K274100 K274100]) downstream as a reporter to yield the DNA damage detection device ([http://partsregistry.org/Part:BBa_K602013 K602013]). Transformed E. coli was exposed to UV light and then incubated for 2 hours. Lycopene as a reporter was extracted from cells with acetone. For details check the Protocols page.
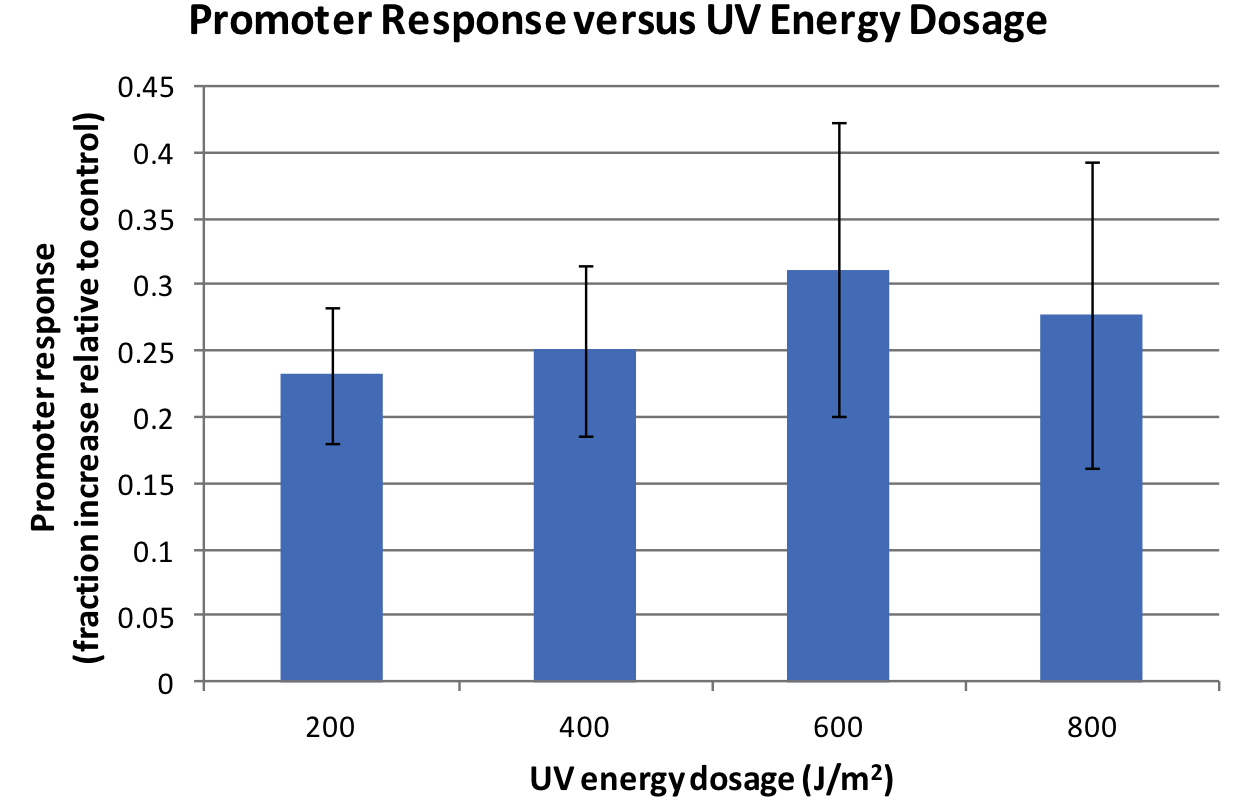
Response was defined as absorbance at 474nm (peak absorbance for lycopene) divided by OD600, followed by subtraction of background (non-irradiated samples) absorbance values. We observed a response to UV irradiation that increased with energy dosage from 200 to 600 J/m^2. Response was decreased at 800 J/m^2, perhaps as a result of intensive DNA damage rendering lycopene biosynthesis genes non-functional.
Work in Progress
DNA damage tolerance
- We are working on assembling a device with all four tolerance genes (PprI, PprA, PprM, RecA) but will not have time to characterize it properly before the wiki freeze. Stay tuned for our poster/presentation at the iGEM World Championship Jamboree for the results!
- To more properly measure the tolerance conferred by each part against DNA double strand breaks (the primary effect of ionizing radiation), we are working on characterizing the parts' tolerances against the drug Mitomycin C. Again, results will be too late for the wiki freeze so catch our presentation at the World Championship Jamboree!
Damage detection
- We have assembled a device utilizing GFP as reporter, but did not have time to characterize it properly. The results will be in our final presentation.
 "
"