Team:Osaka/Tests
From 2011.igem.org
(→SOS promoter assay) |
(→SOS promoter assay) |
||
| Line 21: | Line 21: | ||
<p>We assayed the RecA promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]) by attaching a lycopene biosynthesis gene cluster ([http://partsregistry.org/Part:BBa_K274100 K274100]) downstream as a reporter to yield the DNA damage detection device ([http://partsregistry.org/Part:BBa_K602013 K602013]). | <p>We assayed the RecA promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]) by attaching a lycopene biosynthesis gene cluster ([http://partsregistry.org/Part:BBa_K274100 K274100]) downstream as a reporter to yield the DNA damage detection device ([http://partsregistry.org/Part:BBa_K602013 K602013]). | ||
Transformed ''E. coli'' was exposed to UV light, and then, incubated for 2 hours. Lycopene as a reporter was extracted from cells with acetone. Please check [https://2011.igem.org/Team:Osaka/Protocols Protocol] for details.</p> | Transformed ''E. coli'' was exposed to UV light, and then, incubated for 2 hours. Lycopene as a reporter was extracted from cells with acetone. Please check [https://2011.igem.org/Team:Osaka/Protocols Protocol] for details.</p> | ||
| - | [[File:2011_osaka_promoter_1.png]] | + | [[File:2011_osaka_promoter_1.png|400px]] |
| - | [[File:2011_osaka_promoter_2.png]] | + | [[File:2011_osaka_promoter_2.png|400px]] |
<p>Response is defined as absorbance at 474nm (peak absorbance for lycopene) divided by OD600, followed by subtraction of background (non-irradiated samples) absorbance values.</p> | <p>Response is defined as absorbance at 474nm (peak absorbance for lycopene) divided by OD600, followed by subtraction of background (non-irradiated samples) absorbance values.</p> | ||
Revision as of 19:09, 28 October 2011
Tests
Cell viability
We performed the UV assay. The cells were plated on respective agar plates at different dilutions, air dried, and then exposed to different doses of UV radiation. Plates were wrapped with aluminum foil and incubated in the dark. Colony-forming units were scored after 16h incubation at 37°C. Please check Protocol for details.
PprM gene could also confer high tolerance to inserted cells.
We expected that all cells inserted each genes could increase its ratio of cell survival, however, two genes, pprI and pprA, couldn't confer tolerance. PprI protein is known as a inducer to genes expression such as recA and pprA. Therefore, expression of only pprI may be ineffective for cell survival. Moreover, inserting heavy gene often causes decline of cell survival.
PprA protein has a function for repairing DNA damaged with blunt end. UV exposure causes thymine dimer, not related to blunt end. We suggest that pprA gene may have no function for repairing DNA damaged by UV but some repairing function for other types of damage such as by chemicals, of cause, radiation.
Fortunately, our result about gene mix(connected each genes) showed high tolerance to UV exposure.
RecA gene could induce high cell viability. RecA protein has key role of SOS response. This result revealed that the cell inserted recA gene can get tolerance against DNA damage.
SOS promoter assay
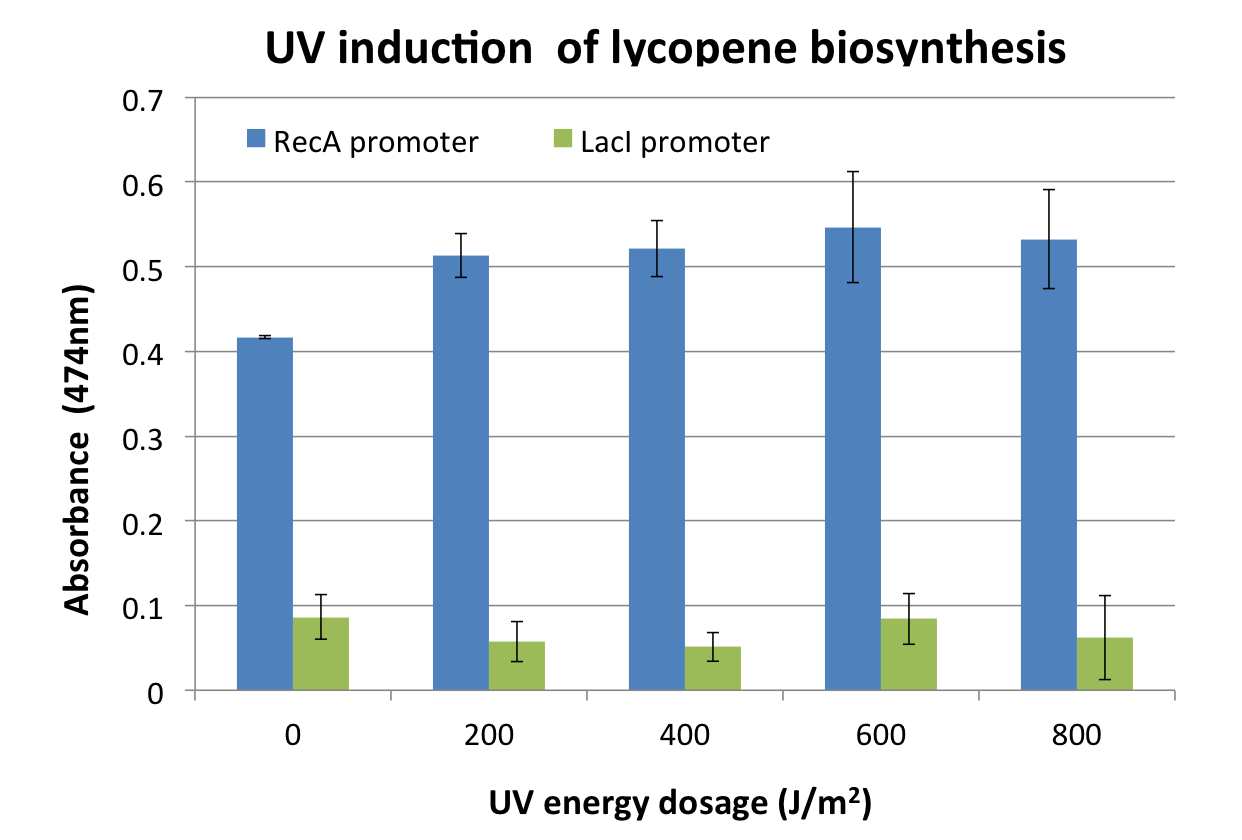
We assayed the RecA promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_J22106 J22106]) by attaching a lycopene biosynthesis gene cluster ([http://partsregistry.org/Part:BBa_K274100 K274100]) downstream as a reporter to yield the DNA damage detection device ([http://partsregistry.org/Part:BBa_K602013 K602013]). Transformed E. coli was exposed to UV light, and then, incubated for 2 hours. Lycopene as a reporter was extracted from cells with acetone. Please check Protocol for details.
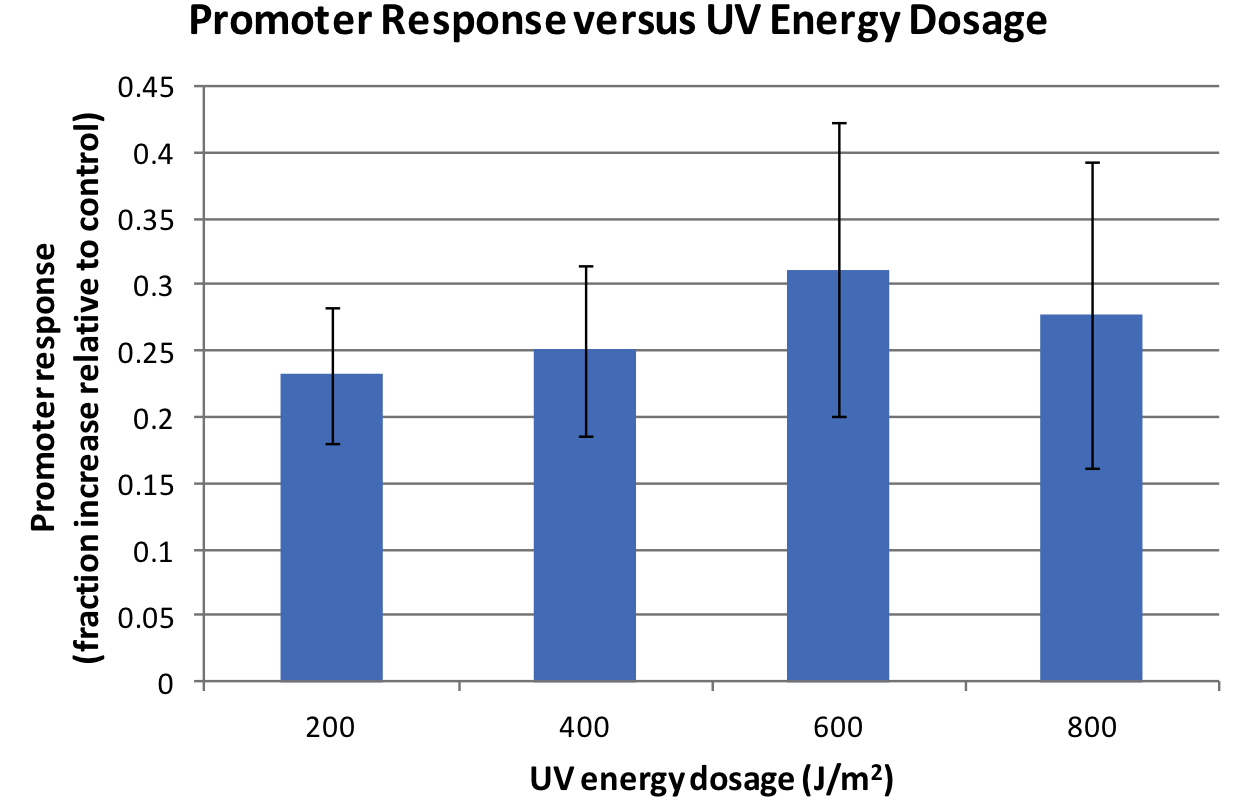
Response is defined as absorbance at 474nm (peak absorbance for lycopene) divided by OD600, followed by subtraction of background (non-irradiated samples) absorbance values.
We observed a response to UV irradiation that increased with energy dosage from 200 to 400 J/m^2. Response decreased with higher energy dosages, perhaps as a result of intensive DNA damage rendering lycopene biosynthesis genes non-functional.
Future work
Cell viability
We created some parts (PprI , PprA , PprM , RecA) but did not have enough time to evaluate them completely.
The effects of each parts to cell viability are shown above, but we will be able to evaluate them more precisely by measuring the effects of other combined parts. (We have already assayed a combined part, mix(all four parts combined).)
We should test responses of cells to other damage types, too.
SOS response
Our project is "Bio-dosimeter", so we should construct two devices about damage tolerance and damage detection respectively.
Our constructed bio-dosimeter shows the level of radiation by one pigment, but it's somewhat difficult to identify the level from light and shade of one color.Therefore, in future, Bio-dosimeter should be going to show the level of radiation using several pigments and promoters.
References
- 放射線抵抗性細菌の新規DNA修復促進タンパク質 , 佐藤勝也 その他 (2006)(Japanese books)
- PprA: a protein implicated in radioresistance of Deinococcus radiodurans stimulates catalase activity in Escherichia coli, Swathi Kota et al (2006)
 "
"