Team:TU Munich/lab/notebook/part4
From 2011.igem.org
(Created page with "<html> <script src="https://2011.igem.org/Team:TU_Munich/slimbox2.js?action=raw&ctype=text/js" type="text/javascript"></script> <link rel="stylesheet" href="https://2011.igem.org/T...") |
|||
| Line 612: | Line 612: | ||
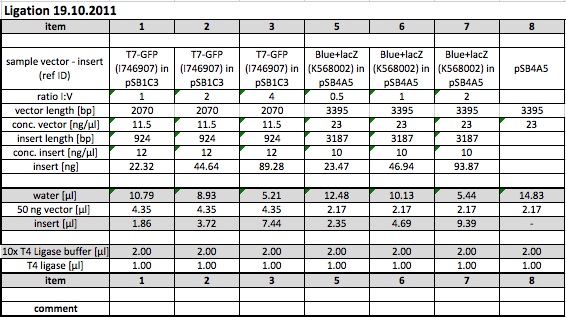
<h2><span class="mw-headline" id="19-10-2011">19-10-2011</span></h2> | <h2><span class="mw-headline" id="19-10-2011">19-10-2011</span></h2> | ||
<p><b>People: Thorsten, Susan, Marta, Leah, Bea, Wolfgang</b></p> | <p><b>People: Thorsten, Susan, Marta, Leah, Bea, Wolfgang</b></p> | ||
| - | <h3><span class="mw-headline" id=" | + | <h3><span class="mw-headline" id="Other-Work">Other Work</span></h3> |
| + | <div class="otherwork"> | ||
<p><i>Picking</i></p> | <p><i>Picking</i></p> | ||
<p>-K568003 in BL21DE3: Plate from 18.10.11 Trafo T7-LacZ clone 2 in BL21 (amp)</p> | <p>-K568003 in BL21DE3: Plate from 18.10.11 Trafo T7-LacZ clone 2 in BL21 (amp)</p> | ||
Revision as of 09:25, 28 October 2011
Cloning Part IV + Results
People: Simon, Alex, Marta, Wolfgang, Bea, Leah, Thorsten, Anna, Susan, Fabi
23-09-2011
People: Simon
Cloning
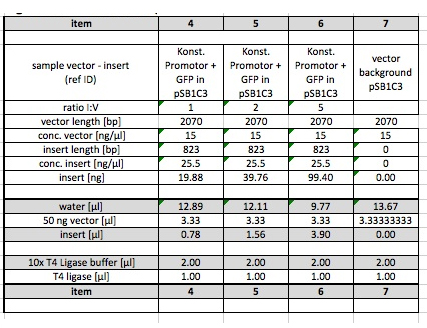
The product of the Ligation from 20-09-2011 (three samples of pSB6A1 with I746907, different I:V-ratios; pSB6A1 E,P cut as vector background) was purified today using glycogen/ethanol precipitation according to protocol.
The yielded product of this was then electroporated into DH5alpha. After incubation at 37 °C for 1 h in 1 ml SOC, 100 µl of the transformed cells were plated onto LB-Amp plates. The remaining 900 µl were spun down (4000 rpm, 5 min), resuspended in 400 µl and 100 µl of this concentrated bacteria culture were again plated onto LB-Agar plates.
24-09-2011
People: Susan
Cloning
The plates 1-3 (containing pSB6A1 with I746907) on which concentrated bacteria soulution was spread all showed lawns and were discarded. The plates 1-3 (containing pSB6A1 with I746907) on which 100 µl of the transformed cells were spread showed >1000 colonies. Plate 4 (vector background) spread with concentrated bacteria solution showed 85 colonies. Plate 4 (vector background) spread with normal bacteria solution showed 24 colonies.
From each of the plates 1 - 3, 4 colonies were picked and inoculated in LB Amp. Incubation at 37 °C over night.
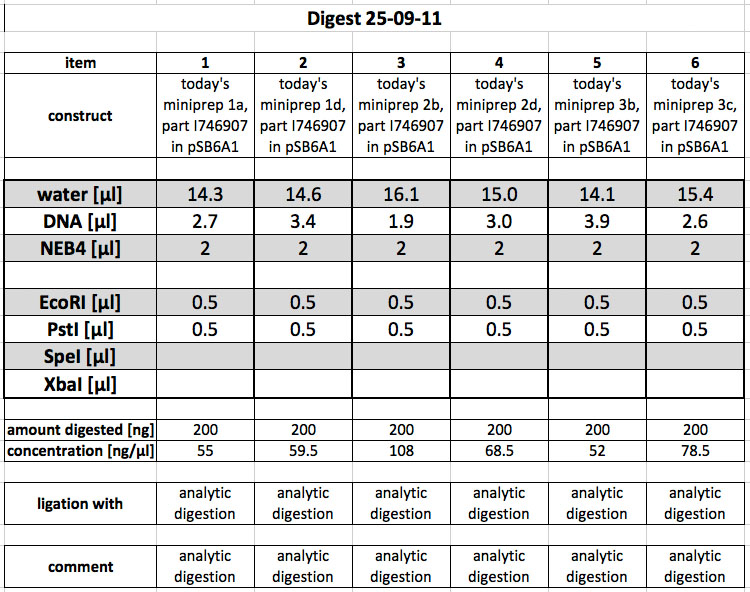
25-09-2011
People: Simon
Cloning
The following overnight samples were miniprepped (all contain pSB6A1 with I746907):
1a, c = 55.0 ng/µl 1d, c = 59.5 ng/µl 2b, c = 108 ng/µl 2d, c = 68.5 ng/µl 3b, c = 52.0 ng/µl 3c, c = 78.5 ng/µl
Digest & Analytic Gel
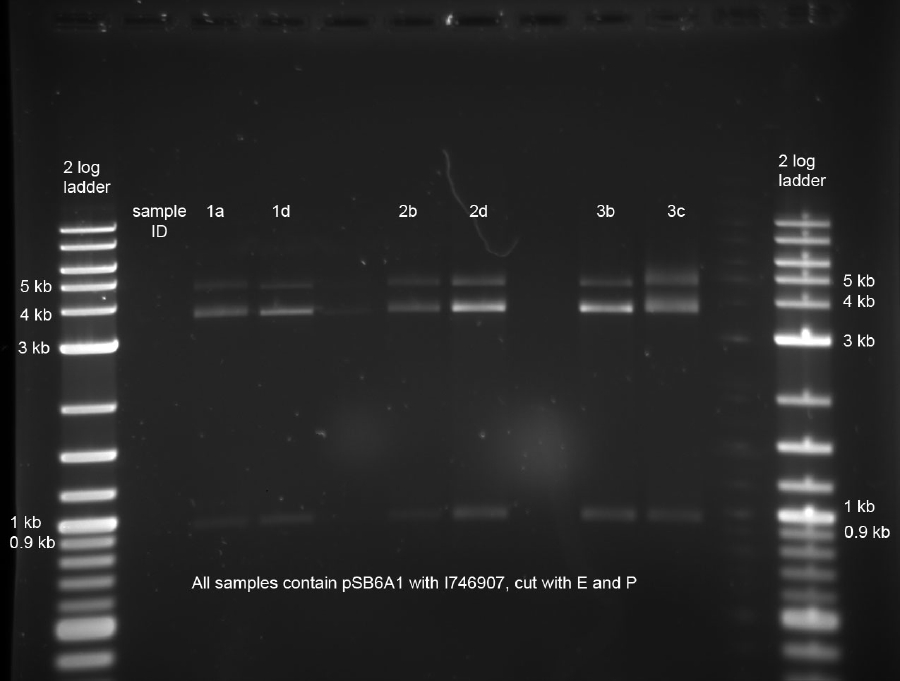
The miniprepped samples were cut using E,P for analytic purposes:
The samples were then separated on an analytical 0.8 % agarose gel:
All bands look fine, with bands around 4 kb (indicating the pSB6A1 plasmids) and 900 bp (indicating the I746907 insert). The band at around 5 kb is uncut vector with insert. This is due to the fact that the digest was incomplete because incubation time was only 10 minutes. However, as can be seen from the gel picture, the HF versions of the restriction enzymes which were used here produce usable results for analytical purposes in that time.
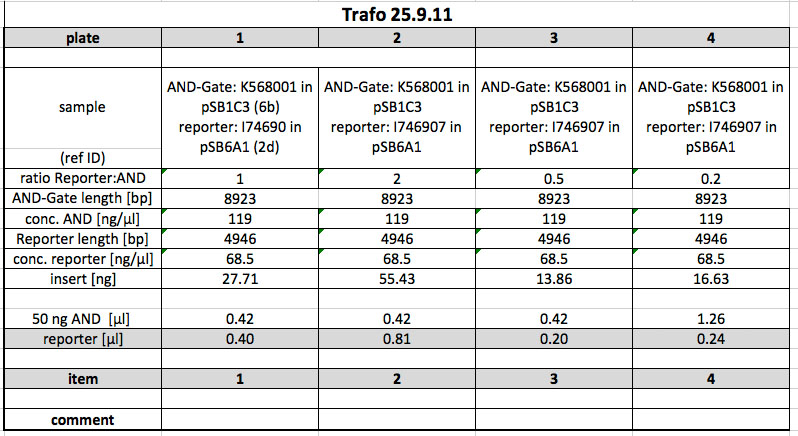
Transformation
Next, both the AND-Gate (K568001, in pSB1C3) and the reporter (I746907 in pSB6A1) were electroporated into CP919. To obtain cells with both plasmids, both plasmids were mixed in different ratios before transformation:
After electroporation and incubation in SOC-medium (1h, 37 °C), the cells were plated onto LB Cam, Amp plates and incubated over night.
26-09-11
People: Marta, Simon
Cloning
Plates 1-3 showed no colonies.
Plate 4 showed 7 colonies. All of them were picked and incubated in 10 ml LB Amp, Cam at 37 °C over night.
27-09-11
People: Marta, Simon
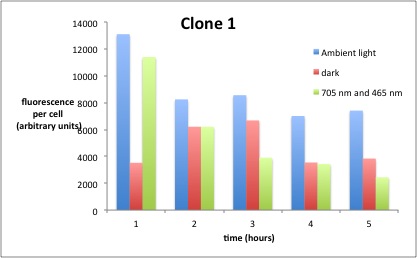
Testing
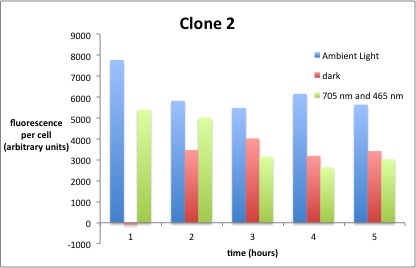
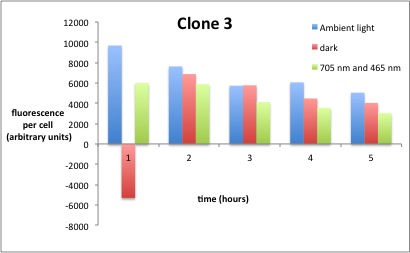
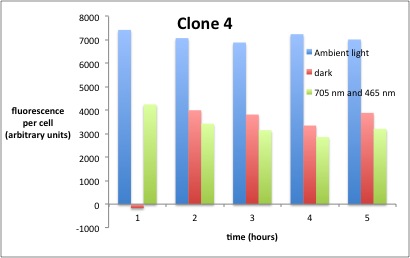
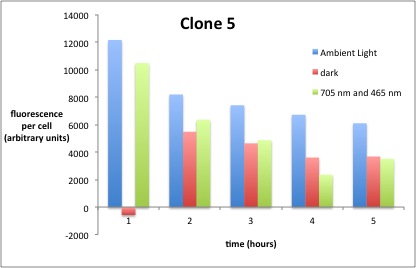
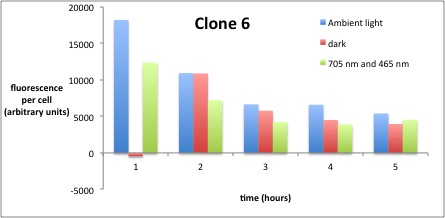
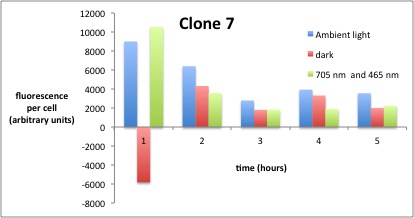
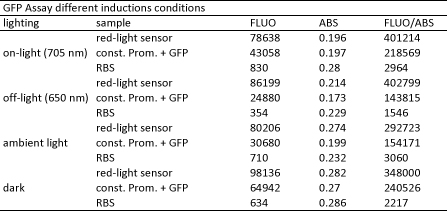
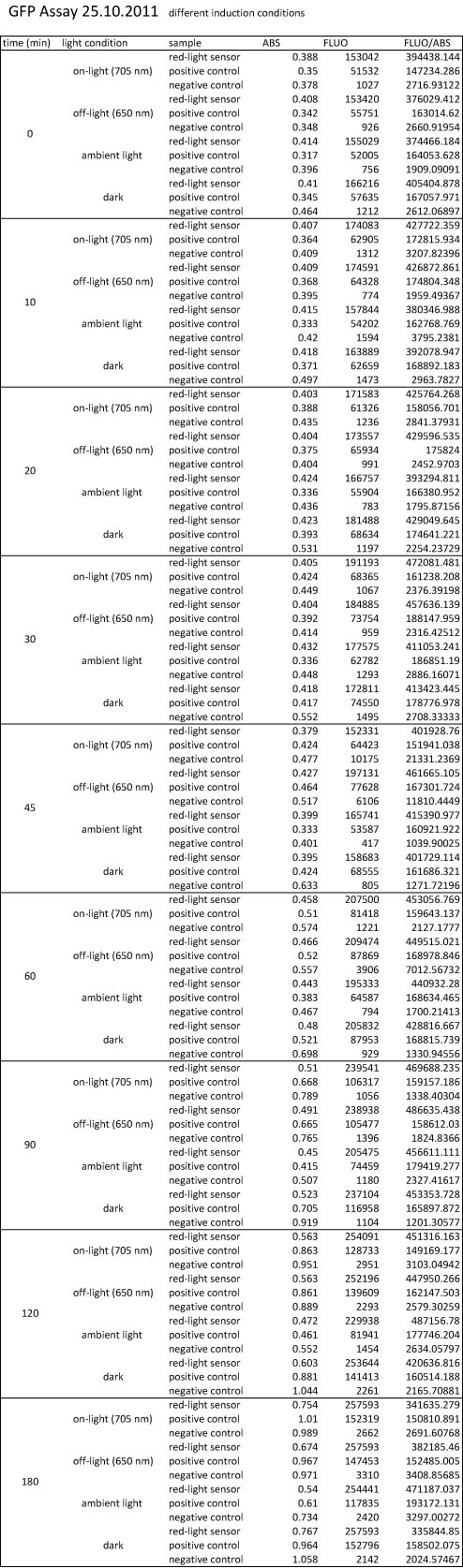
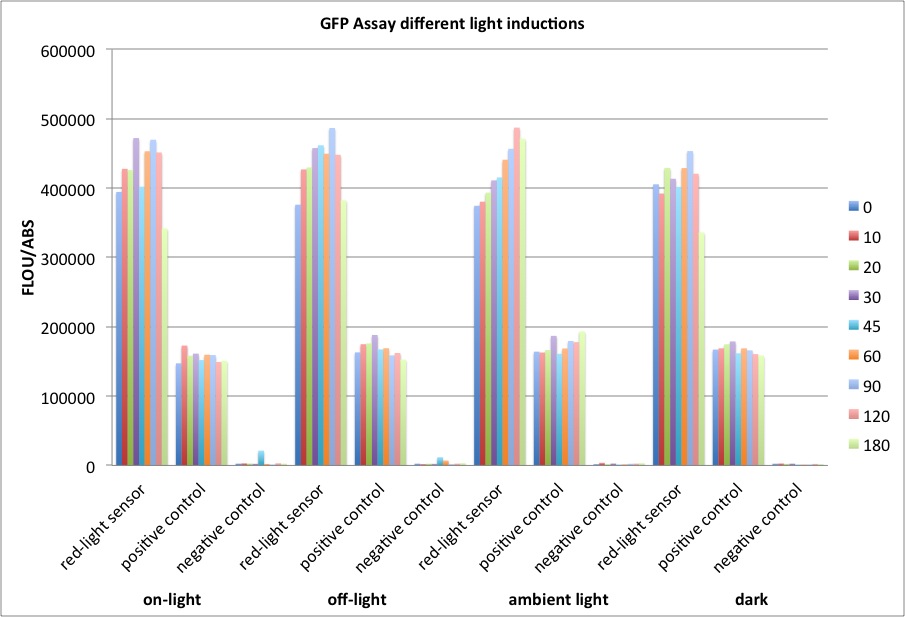
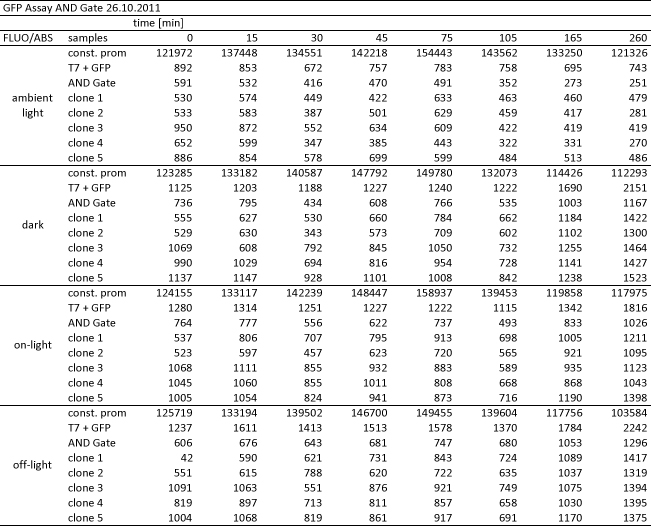
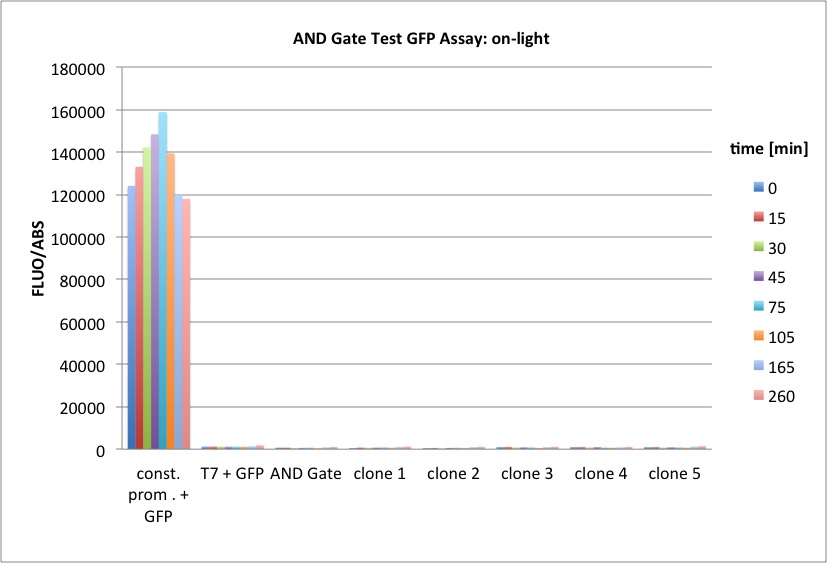
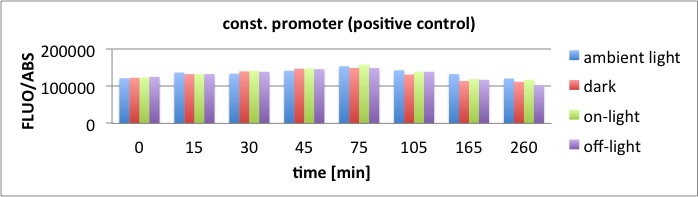
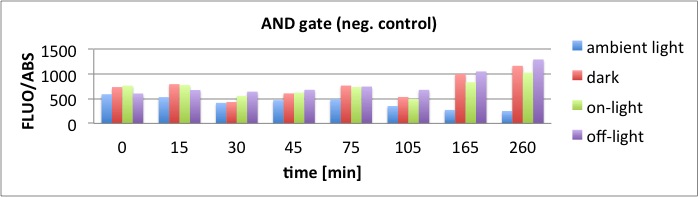
We performed GFP-Assays with all 7 overnight cultures. All cultures were diluted to an OD of approx 0.2. Then, the GFP fluorescence and the OD600 was measured every hour. Also, a blank of LB medium was measured every hour (both fluorescence and OD600). Between measurements, the cells were incubated under different lighting conditions at 37 °C:
- Blue light (465 nm) and red on-light (705 nm)
- ambient light (lamp of the incubator)
- in the dark
The fluorescence per cell can be calculated using the following formula:
"Fluorescence per cell" = (fluoresence(sample) - fluorescence (blank)) / (OD600(sample) - OD600 (blank))
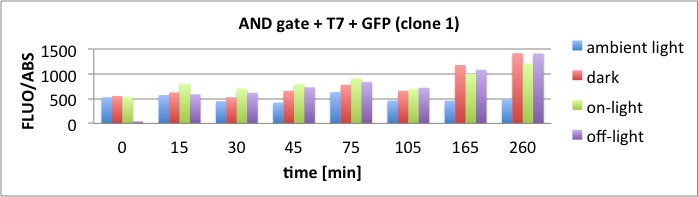
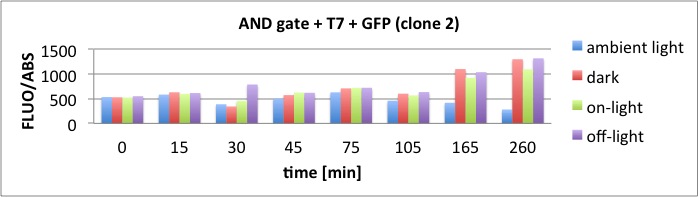
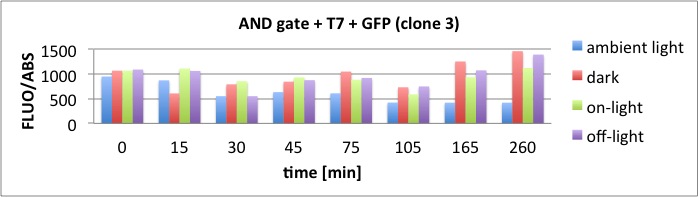
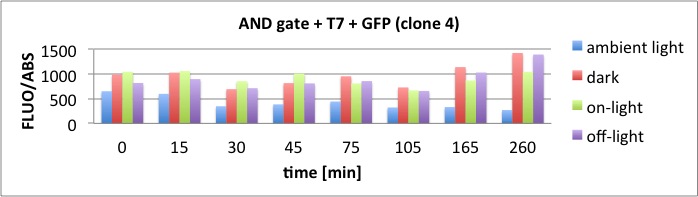
Results
The results of the GFP-Assay are shown here:
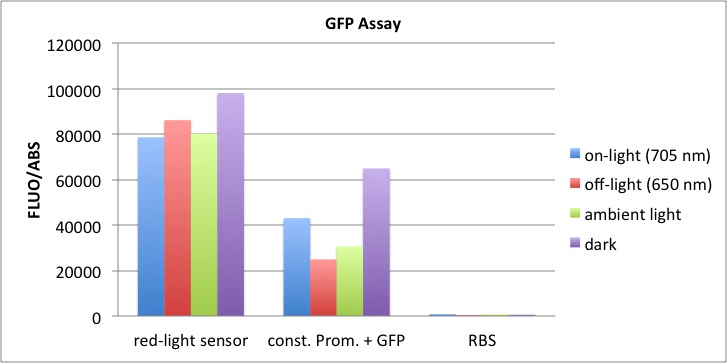
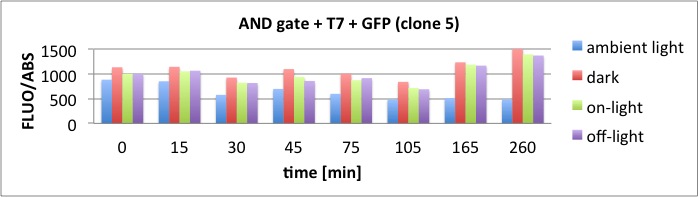
At t = 1h, the data is as expected: high fluorescence in the ambient light and the red & blue light samples, very low fluorescence in the dark samples.
Then, over the next few hours, this changes dramatically: the fluorescence per cell decreases.
The diagrams show that light does influence the GFP production in CP919 with our constructs. However, a definite quantitative statement based on this data is not possible. Further testing is needed, especially with regard to the following questions:
- What is the temperature optimum of the blue light sensor and how does this influence the AND-Gate? For this, characterization of the bluelight sensor should be considered (see other iGEM teams).
- Does the red light sensor work correctly (which could not be proven by previous experiments)? As no other teams managed to get this part to work properly, we need to characterize it further. The best way to do this would be testing of the following construct: BBa_K568000 + RBS + GFP
- Has the red ligth sensor an asymptotic on steady state such that further radiation with red off light is needed ?
- What are the optimum and maximum durations of measurement? For this, the modeling results should be considered.
06-10-11
People: Alex, Simon, Marta
Cloning
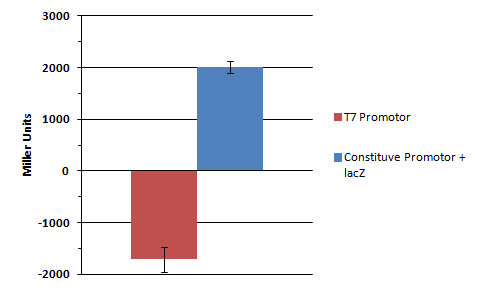
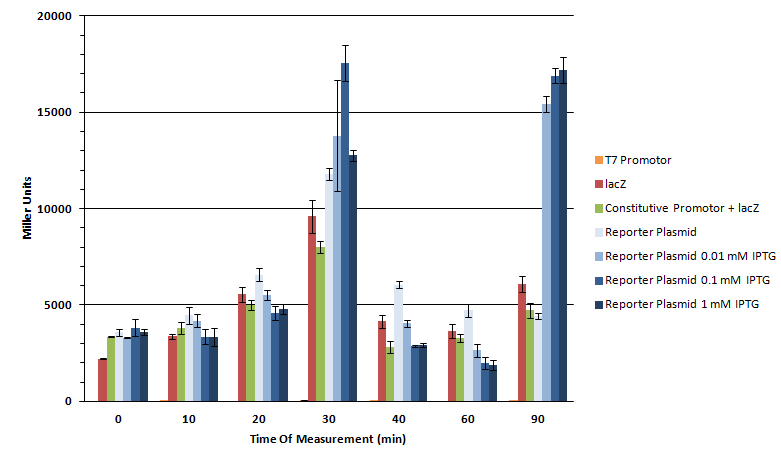
Parts BBa_I712074 (Kan; T7 prom) and BBa_I732017 (Amp; lacZ) were transformed into BL21 cells and incubated over night on the appropriate antibiotics.
Other work
Chemical competent BL21 (DE3) col+ RLF (?) cells were obtained from Lars Mitschke. In order to prepare electrocompetent cells, 20 ml LB medium are inoculated with 1 µl cells and incubated over night.
07-10-11
Alex, Simon, Marta
Cloning
New parts
The parts BBa_J23100 (Constitutive promoter), BBa_PSB4A5 (low copy plasmid) and BBa_K081012 (rbs-GFP) were transformed from the 2011 DNA distribution kit plates into DH5alpha using different amounts of DNA. This way we obtained 3 sample for each part with the following volumes of DNA:
- Sample 1 - 0.5 µl
- Sample 2 - 1.0 µl
- Sample 3 - 8.0 µl
The RBS (BBa_J44001)was also transformed into DH5alpha. In this case we only had one sample with 0.5 µl DNA.
The 10 obtained cell suspensions were spread on agar plates and incubated overnight.
Results
rbs and lacZ in BL21
The transformation of rbs into BL21 was successful. 3x 5 ml were inoculated with clones and incubated over night. The transformation of lacZ did not yield any colonies.
Other work
We prepared some electro-competent cells. For this purpose we used BL21 (DE3) col+ rlf, which we had obtained from prof. Buchner the day before.
08-10-11
People: Alex
Cloning
T7 prom
A glycerin stock was prepared using the BL21 (DE3) cells transformed with T7 prom from yesterday.
Transformations
LB was inoculated with the cultures, using the following antibiotics: BBa_K081012 (3 clones used; Kan), BBa_J44001 (1 clone used; Amp), BBa_J23100 (3 clones used; Amp) and pSB4A5 (3 clones used; Amp).
Results
Transformations
All transformations from yesterday yielded colonies. The colonies from BBa_J23100 (const. prom) and from pSB4A5 were red (RFP!).
Contamination test of new electrocompetent cells
Amp and Kan plates did not yield any colonies. However, on the Cm plate, a slight, but confluent layer of bacteria could be detected.
09-10-11
Overview of Parts
| Part | Function | To be digested with (on Monday) | Plasmid-Backbone |
|---|---|---|---|
| BBa_J23100 | constitutive promotor | S,P | BBa_J61002 (Amp & RFP) |
| BBa_I732017 | lacZ | X,P | pSB1A2 |
| BBa_J44001 | rbs | S,P | pSB1A2 |
| BBa_K081012 | rbs-GFP-term | X,P | pSB1AK3 |
| BBa_I712074 | T7 promotor | S,P | pSB1AK8 |
Cloning
Minipreps
All overnight cultures were miniprepped.
1: J23100 in J61002, c = 89.5 ng/µl
2: J23100 in J61002, c = 61.0 ng/µl
3: J23100 in J61002, c = 84.5 ng / µl
4: pSB4A5, c = 113 ng/µl
5: pSB4A5, c = 127 ng/µl
6: pSB4A5, c = 175 ng/µl
7: K081012 in pSB1AK3, c = 79.0 ng/µl
8: K081012 in pSB1AK3, c = 114 ng/µl
9: K081012 in pSB1AK3, c = 66.5 ng/µl
10: J44001 in pSB1A2, c = 83.5 ng / µl
Planing of cloning
K568001 in pSB4A5:
- Cut K568001 from pSB1C3 with E, P
- Open pSB4A5 with E, P
- ligate
I746907 in pSB1C3:
not necessary, because we make our own T7prom-GFP construct, because then we have the same GFP in all parts (reporter, positive control, light sensor-GFP constructs...)
Red Light Sensor with GFP:
- Cut K081012 (rbs-GFP-term, in pSB1AK3, prepped 09.10.11) with X, P
- Cut K568000 with P, S
- Ligate (products cut with X and S can be ligated → scar)
'Constitutive Promoter and GFP:
- Cut K081012 (rbs-GFP-term) with X, P
- Cut J23100 (const. prom in vector J61002) with S, P
- ligation
This results in J23100-K081012 construct in pSB1A2 (see description of vector J61002)
Constitutive Promoter and lacZ:
- Cut J23100 (const. prom in vector J61002) with S, P
- Cut I732017 (lacZ) with X, P
- ligation
This results in J23100-I732017 construct in pSB1A2 (see description of vector J61002)
T7promoter and GFP:
- Cut I712074 (T7 prom, in pSB1AK8) with S, P
- Cut K081012 (rbs-GFP-term) with X, P
- Ligate
For re-characterization of blue light promoter (blue light prom + lacZ):
- Part K238015 could be used for this (this part is the blue light promoter plus GFP E0040, which is the same version of GFP as in K081012), but is not on a shipment plate → need to make that part.
- → ligate blue light promotor with K081012
- use the tube labeled „8, blue light prom, cut with S, P, 1.9.“ (is in „Wichtige Dinge Box“)
- cut K081012 with X, P
- ligate
- vector: pSB1A2 (according to wiki. It doesn’t say anything on the tube, and because there are only a few µl left, after ligation and trafo the cells should be plated on three plates (with amp, kan or cam) to make sure we definately have something!)
Other Work
LB-Agar plates with Amp, Cam, or Kan were prepared.
Some 1.5 ml and 2 ml Eppendorf tubes were autoclaved.
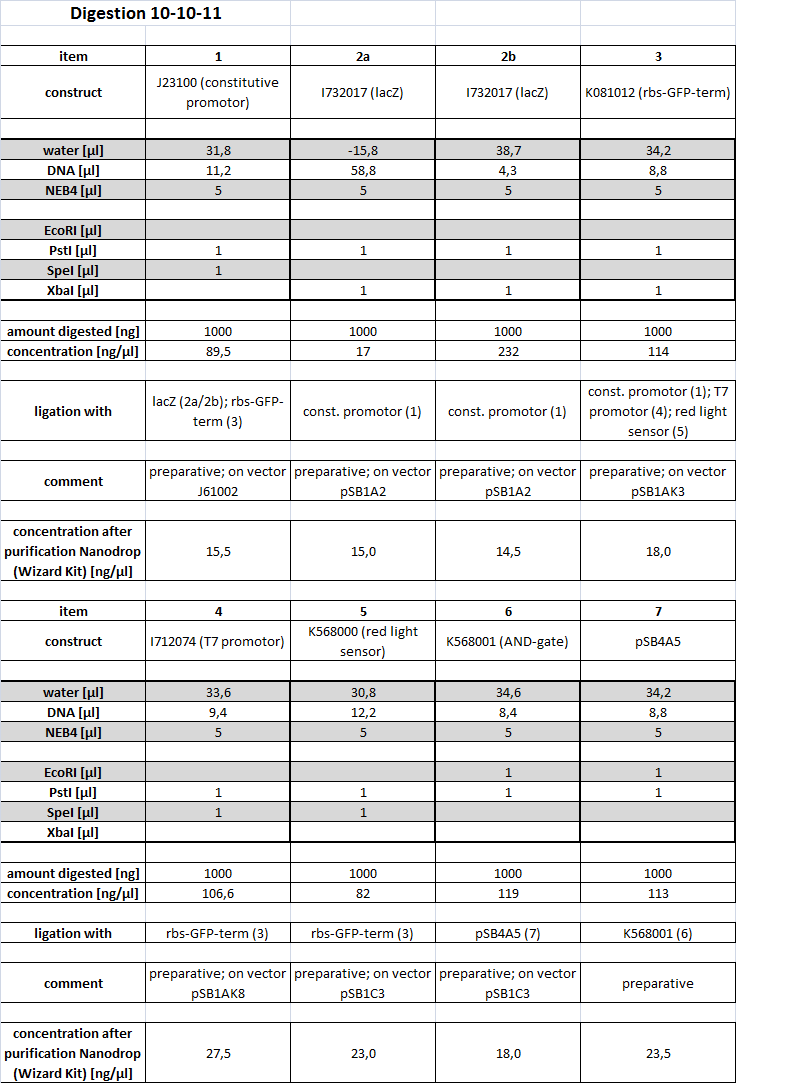
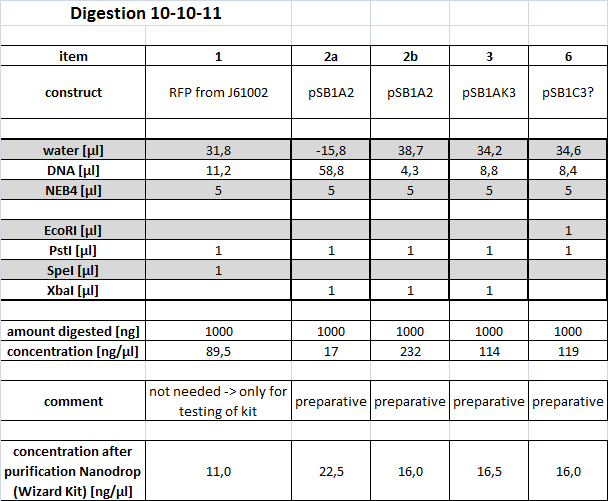
10-10-11
People: Alex, Marta, Simon
Cloning
Restriction (IDA1)
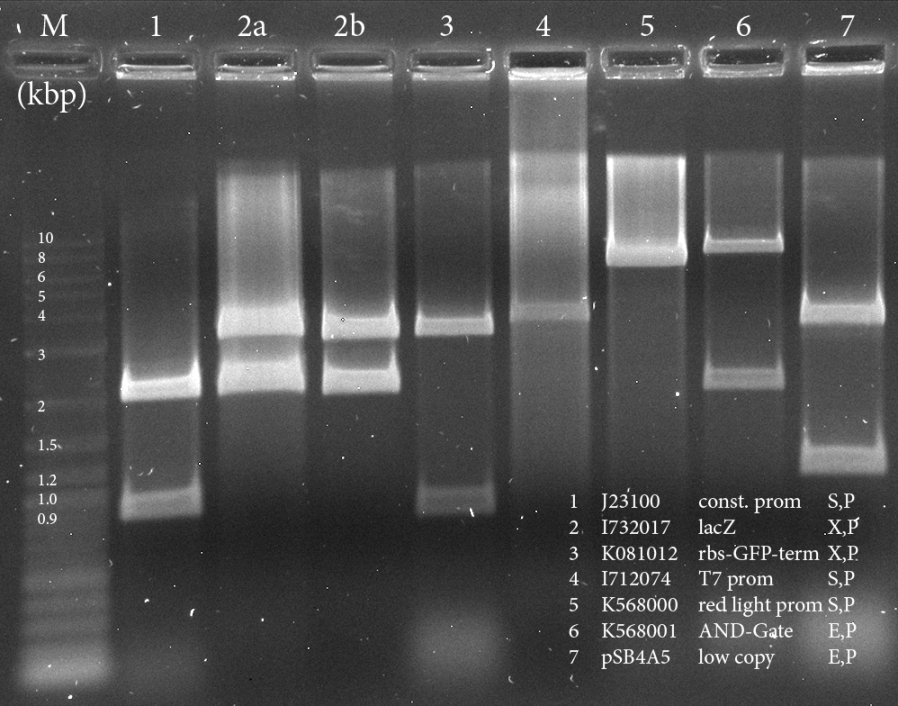
We needed to digest existing parts in order to be able to construct the following parts: - constitutive promotor with lacZ - constitutive promotor with rbs-GFP-term - T7 promotor with rbs-GFP-term - blue light sensor with rbs-GFP-term - red light sensor with rbs-GFP-term - AND-gate in pSB4A5 (low copy vector)
All tubes of this Digest are stored in "Wichtige Dinge" Box 4 and marked with AH2.
All tubes of the this Digest are stored in iGEM Box 3 and marked with IDA1.
The samples were applied onto a 0.8 % Agarose gel. We expected the following sizes of fragments:
| Part | Insert | Backbone (+ Insert) | Restriction |
|---|---|---|---|
| 1: BBa_J23100 | 0.8 kbp | 2 kbp | S,P |
| 2: BBa_I732017 | 3 kbp | 2 kbp | X,P |
| 3: BBa_K081012 | 0.8 kbp | 3.2 kbp | X,P |
| 4: BBa_I712074 | - | 3.5 kbp | S,P |
| 5: BBa_K568000 | - | 5.8 kbp | S,P |
| 6: BBa_K568001 | 6.8 kbp | 2 kbp | E,P |
| 7: pSB4A5 | - | 4.5 kbp | E,P |
The parts written in orange are needed for the following ligations. The others are extracted as well, in order to obtain part backups. The gel was run for 1:30 h at 110 V.
All the bands are as expected and were extracted using the kit from Qiagen, as the Wizard kit was empty.
Transformation
Transformations were prepared of the parts used in digestions 2a, 2b, 4 and 5, in order to have DNA-backup clones.
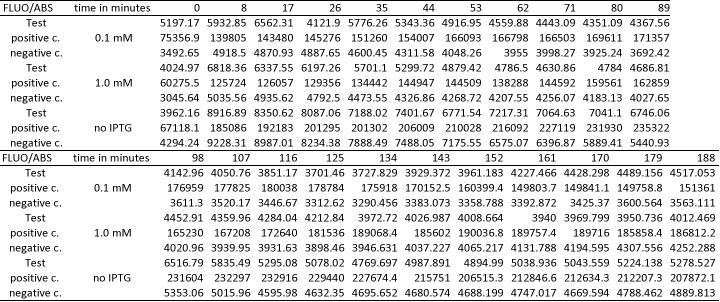
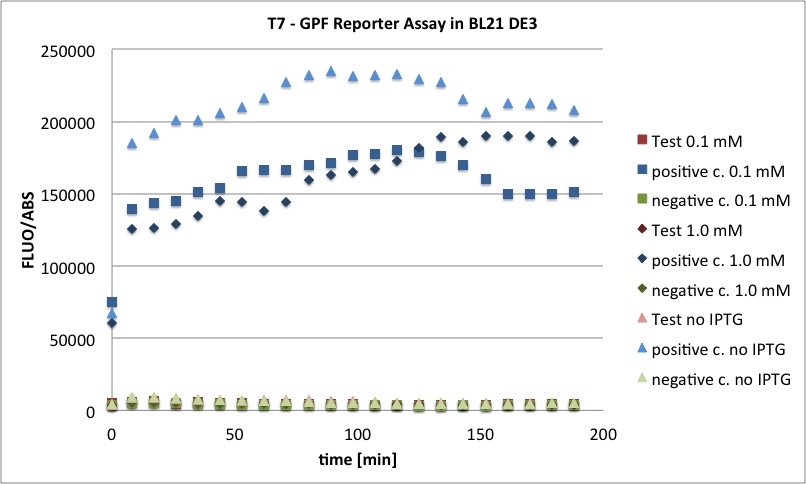
Testing
We want to redo the characterization of AND-Gate with I746907. The liquid cultures of the CP919 cells with which the characterisation was performed on 27-09-11 were stored in the fridge since then. Today, 50 µl of the cultures of trafo 4, clones 4 and 6 were spread onto LB Cam, Amp plates to see if the bacteria survived the 2 weeks in the fridge. If there are colonies, they should be inoculated and used for testing again. In addition to this, glycerol stocks should be prepared from these cells.
11-10-11
People: Alex, Simon, Marta
Testing
Testing of AND-Gate in CP919
From each of the two plates with CP919 containing K568001 and I746907, 2 colonies were picked and inoculated into 10 ml LB Amp, Cam, and incubated at 37 °C.
The original plan was to splut the cultures into 3 parts once the bacteria had grown to an OD600 of 0.1 and then continue incubation as follows:
- Incubation in the dark, 37 °C
- Incubation under red off-light (650 nm), 37 °C
- Incubation under ambient light, 37 °C
However, the cultures grew very slowly if at all. Maybe they didn't keep both AB resistances during storage in the fridge and the reason why we got colonies on the LB Amp Cam plates is that the plates were too old (23-09-2011). Because of the D600 was approx 0.006 after a few hours of incubation, this might be the case. Because of that, the cultures were not split as mentionned above. Instead, they were incubated under red-off light at 37 °C over night.
Cloning
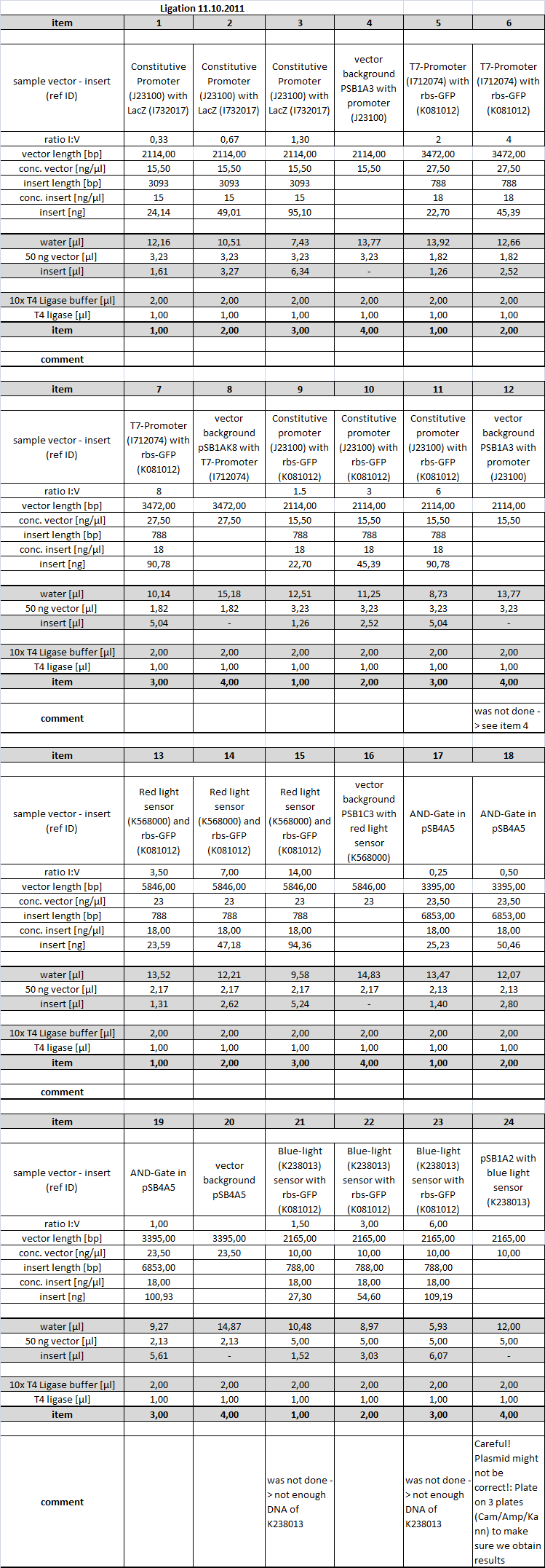
The restricted parts from yesterday were ligated according to the following scheme:
Ligations 12, 21 and 23 were not performed.
Other Work
In order to further test the prepared electrocompetent BL21 (DE3), bacteria, which grew on the Cm-Plate were inoculated into LB (Cm) and incubated over night.
Results
Both plates on which the CP919 cells with Andgate and reporter were spread yesterday showed many colonies.
12-10-11
People: Marta, Alex
Cloning
Transformations
The following transformations were performed:
- I732017 (lacZ) in BL21 (DE3) -> negative control for Miller Assay; plated onto Amp
- I712074 (T7 promotor) in BL21 (DE3) -> negative control for Miller Assay; plated onto Kan
- K238013 in DH5 alpha -> DNA backup; plated onto Amp and Cm
Other Work
A new chloramphenicol stock (30 mg/ml in 100 % ethanol; sterile filtrated) was prepared.
Results
Contamination test BL21 cells:
The cells grew in the LB (Cm) medium. The strain we obtained has a natural chloramphenicol resistence. Thus, the growth is expected.
13-10-11
People: Marta
Cloning
"Transformations"
The samples from the following Ligations were Transformed again and plated on agar:
- Ligation 5
- Ligation 7
- Ligation 13
- Ligation 15
Also the Ligations 6 and 14 were plated again using a gretaer volume (200µl instead of 150 µl)
Colonies from the ligations that worked have been picked and will be incubated over night in liquid agar.
Results
These are the results of the Ligations that were plated the day before and cultivated over night.
| Ligation number | Colonies? | Comments |
|---|---|---|
| 2 | yes | |
| 4 | yes | |
| 6 | no | Transformed cells were plated again and a new transformation was carried out. |
| 8 | no | |
| 10 | yes | |
| 14 | no | Transformed cells were plated again and a new transformation was carried out. |
| 16 | no | |
| 18 | yes | |
| 20 | yes | |
| 22 | yes | |
| 24 | yes | |
| LacZ in BL21 | yes | |
| T7 in BL21 | yes | Picking was not possible, so a dilution was plated on agar. |
| K238013 in DH5alpha (Amp) | yes | |
| K238013 in DH5alpha (Cm) | yes | Colonies are violet |
14-10-11
People: Alex, Simon, Marta, Bea, Thorsten, Fabipups
Cloning
Digestion
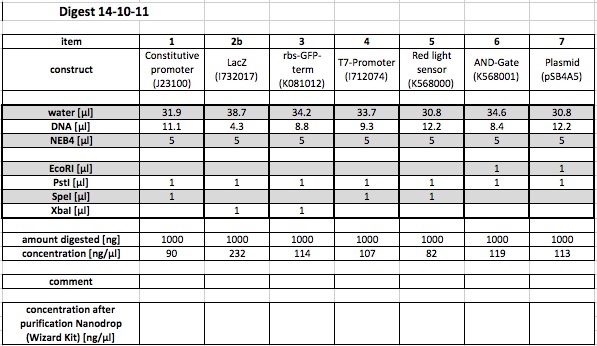
Digestion of the following parts in order to repeat some of the ligations:
Plasmid Mini-Preps and Gylcerol-Stocks
Over night cultures from 13-10-11 were mini-prepped and glycerol-stocks were made. If no strain ist mentioned, the parts were transformed into DH5 alpha cells. The plasmid mini-preps and the glycerol-stocks are marked "AH4".
| Part | Function | Concentration (if prepped) | Glycerol-stock prepared? |
|---|---|---|---|
| I732017 | lacZ | 138 ng/µl | yes |
| I732017 | lacZ (other charge) | 110 ng/µl | yes |
| I712074 | T7 promotor (clone 1) | 113 ng/µl | no |
| K568000 | red light promotor | 94.5 ng/µl | yes |
| I712074 | T7 promotor | 141 ng/µl | yes |
| K238013 | blue light promotor | 176 ng/µl | yes |
| K238013 | blue light promotor (other charge) | 218 ng/µl | no |
| I712074 (BL21) | T7 promotor | no prep | yes |
| I712074 (BL21) | T7 promotor | no prep | yes |
Inoculation
Due to too much growth, the transformation of I732017 (lacZ) in BL21 was diluted and plated again yesterday. Today, LB (Amp) was inoculated with two different colonies and incubated over night at 37 °C.
Ligation
In order to confirm our results we transformed the following ligations, that had not been transformed yet:
- Ligation 1
- Ligation 3
- Ligation 13
- Ligation 15
Ligation 13 and 15 had already been transformed the day before, but only with a small amount of sample. This time we used the entire volume in order to obtain more colonies.
This week's ligations had been plated with only 150µl of the cells culture. In order to increase the density on the plates we plated the ligations again, this time using the entire volume. The Ligations that we transformed today were also plated this way.
overview
Blue light promoter:
"This part was actually put in a pBR322 vector. This is a vector which has a low copy number (around 15-20). This is needed to make the promoter work properly in E.Coli. The low copy number ensures a good ratio between the promoter and its repressor, which is naturally present in E.Coli. When a high copy number is used, the promoter is abundant which results in a constituitivly active promoter." source http://partsregistry.org/Part:BBa_K238015:Design
--> subcloning of K568003 into low copy vector pSB4A5 (was in pSB1A2)
15-10-11
People: Marta
Cloning
Preparation of restricted DNA and Ligation
The T7 Promoter was cut with S,P and not with E,P as described in the picture.
The different bands were cut out and stored in the -20 freezer in the Box 3. (The tubes are labeled 1-7 and contain pieces of gel)
Results
Ligations
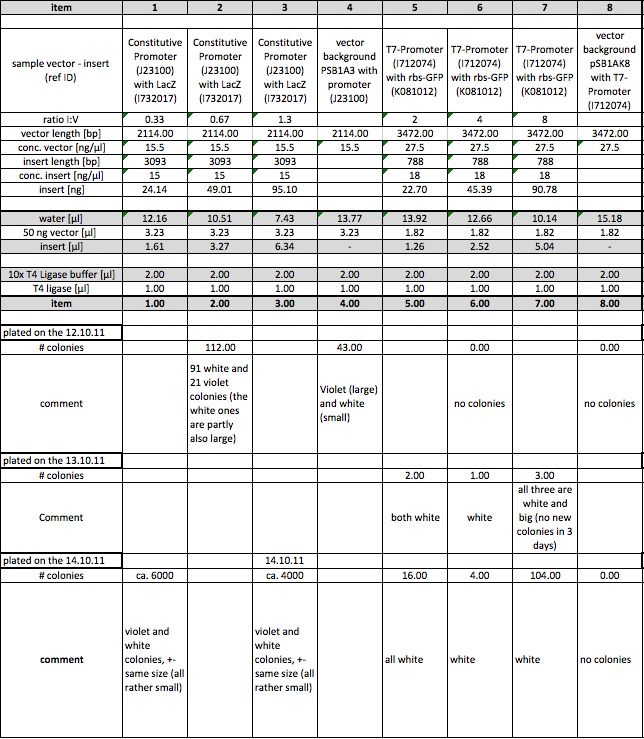
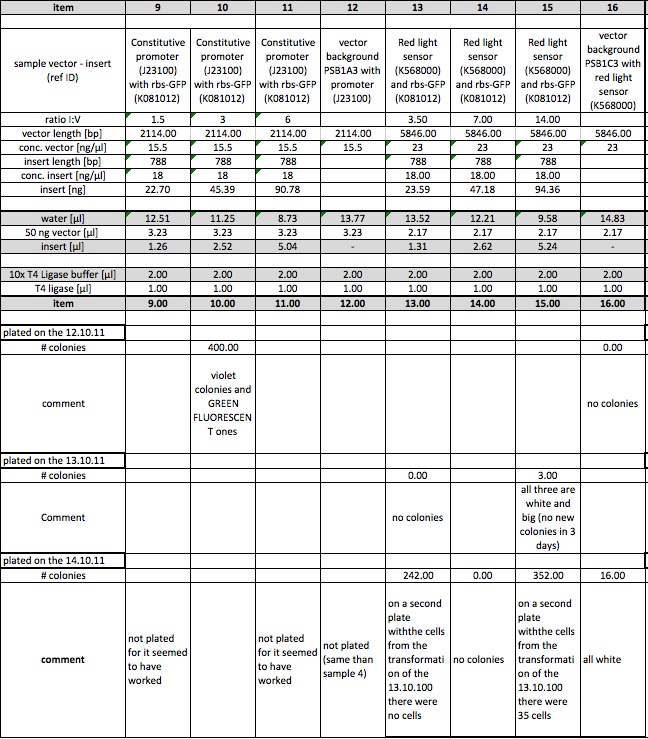
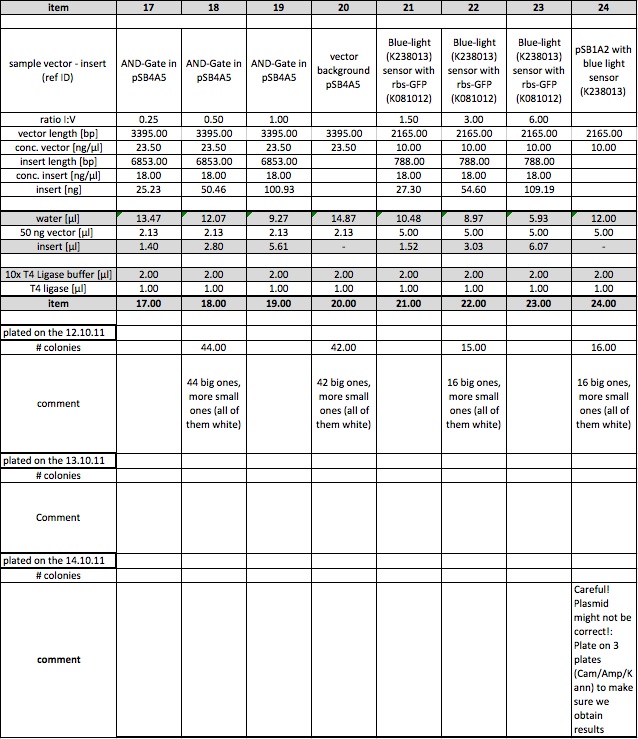
These are the results of our ligations. We plated on three different dates (12.10.2011, 13.10.211 and 14.10.2011) depending on the results.
Constitutive promoter (J23100) with LacZ (I732017)
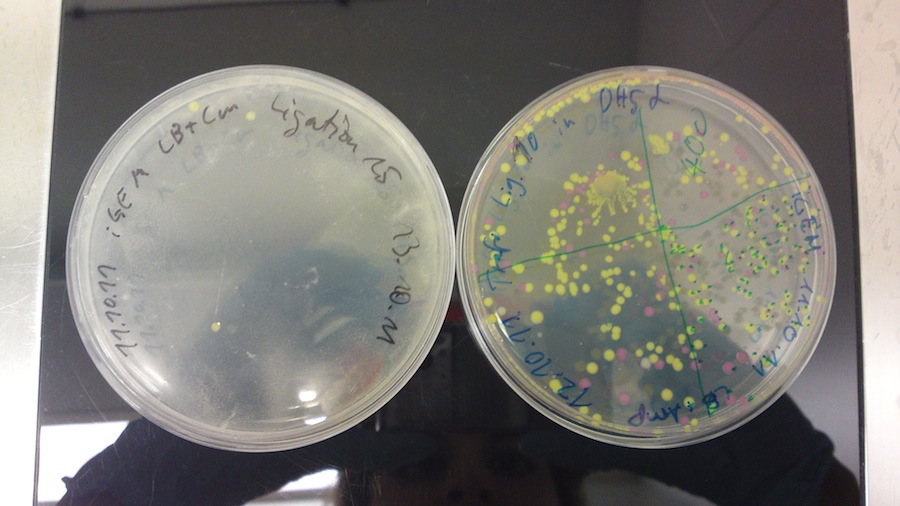
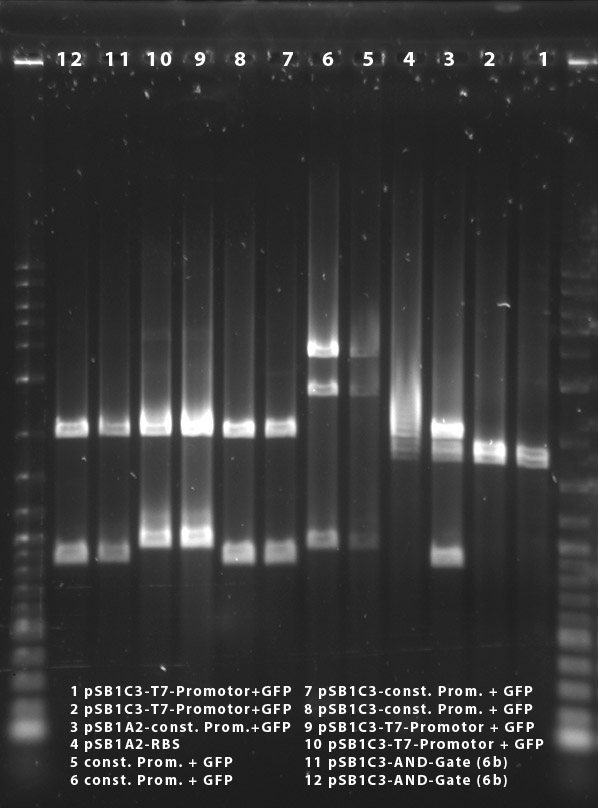
The plasmid contains an RFP-casette that is cut out when the ligation is performed correctly. We could observe, that it takes some time for the cells to produce the RFP. After they have reached a certain size, they turn violet. As the table shows we have both violet and white colonies. In order to obtain the right cells we will have to pick the withe colonies. This ligation probably worked, since the cells of the vector background, that serve as a control are mostly violet. The ones that are white are not big enough to produce the RPF, but are likely to do so after further incubation.
T7-promoter (I712074) with rbs-GFP (K081012)
The measurements from the 12.10 and the 13.10 were performed with a very low concentration, which is probably the reason why we otained almost no cells. The results from the 14.10 prove that the ligation must have worked, for we obtained cells in all samples but the vector background.
Constitutive promoter (J23100) with rbs-GFP (K081012)
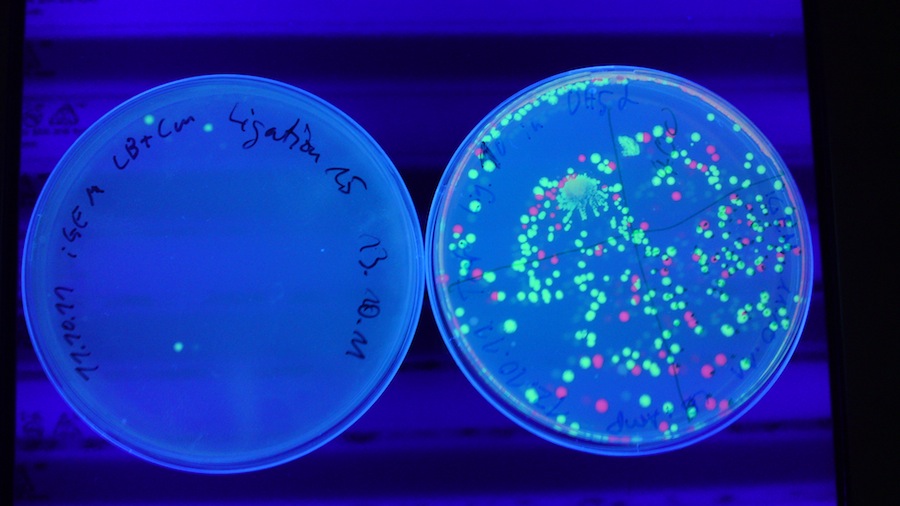
Under the UV-Lamp it is easy to distinguish the cells containing RFP from the ones containing GFP. The fact, that there are green fluorescent cells means the ligation has worked.
Red light sensor (K568000) and rbs-GFP (K081012)
This ligation also seems to have worked. even thought there are some cells on the plate with the vector background, the ration is still ok. Also we could observe fluorescence in the sample number 15. The others didn't show luminiscence yet for they had not been incubating as long as sample 15
AND-Gate (K568001) in psBB4A5
Since there are as many colonies on the ligation-plate than on the vector background we believe this ligation did not work. We have to find out why the cells did not turn violet, as they should have, for the psBB4A5 contains an RFP-casette, that is cut out when the ligation is correctly performed.
Blue light sensor (K238013) with rbs-GFP (K081012)
The data indicates, that the ligation may not have worked. Since we decided to characterize the blue light sensor with lacZ we will probably not repeat this ligation even though it didn't work.
Ligations: Excitation Test
Colonies on ligations 10 (const. promotor with GFP) and 15 (red light sensor with GFP) produced (green to yellow) fluorescence when excited with light of 380 nm. This might imply that the ligations worked and the parts are successfully producing GFP. In the case of red light sensor with GFP, the ambient light seemed to be sufficient to trigger a leaky production of GFP.
17-10-11
Cloning
Ligations from 11-10-11
In order to find out, whether the ligations of const. prom. + lacZ in DH5 alpha were successful (Ligations 1-4), 2 colonies of each ligation plate were inoculated into LB (Amp). On the next day, they will be tested using ONPG and negative controls. Furthermore, 6 colonies of T7 prom. + rbs-GFP (2x Lig. 7 14.10.; 1x Lig. 5 14.10., Lig. 6 14.10., Lig. 7 13.10., Lig. 5 13.10.) were inoculated into LB (Kan), in order to prep the plasmids, if necessary.
Results
18-10-11
People: Wolfgang, Bea, Fabi, Marta, Thorsten, Leah, Susan
Cloning
Purification
The samples (from the digestion) that were put in the freezer on the 15.10.11 were purified with the wizard promega kit.
Following concentrations were measured:
1) const.prom J23100 (P,S) 22,5 ng/µl
2) LacZ I732017 (P,X) 22,0 ng/µl
3) cbs-GPS-term K081012 (P,X) 19,6 ng/µl
4 )T7-prom I712074 (S,P) 21,5 ng/µl
5 )Red light K568000 (P,S) 20,5 ng/µl
6 )AND Gate 568001 (E,P) 14,5 ng/µl
7) Plasmid psB4A5 (E,P) 23 ng/µl
Mini-Prep
Mini-prep (Metabion) of the overnight cultures from the 17.10.11.
1) Lig.7: T7+GFP in DH5alpha (Ligation 13.10.11) 87,5 ng/µl
2) Lig.7: T7+GFP in DH5alpha Kan (Ligation 14.10.11) 52,5 ng/µl
3) Lig.7: T7+GFP in DH5alpha Kan (Ligation 16.10.11) 125,0 ng/µl
4) Lig.5: T7+GFP in DH5alpha Kan (Ligation 14.10.11) 82,5 ng/µl
5) Lig.5: T7+GFP in DH5alpha Kan (Ligation 13.10.11) 113,0 ng/µl
6) Lig.6: T7+GFP in DH5alpha Kan (Ligation 14.10.11) 85,0 ng/µl
7) Lig.2: const.prom+LacZ in DH5alpha Amp (Ligation 12.10.11) 77,5 ng/µl
8) Lig.2: const.prom+LacZ in DH5alpha Amp (Ligation 12.10.11) 82,5 ng/µl
9) Lig.3: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 82,5 ng/µl
10) Lig.3: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 205,0 ng/µl
11) Lig.1: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 90,0 ng/µl
12) Lig.1: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 65,0 ng/µl
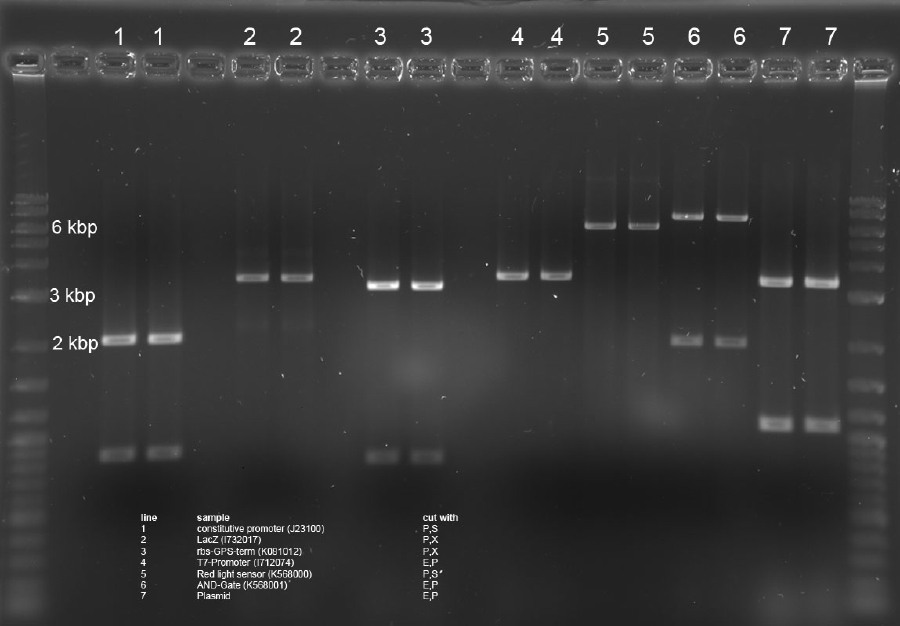
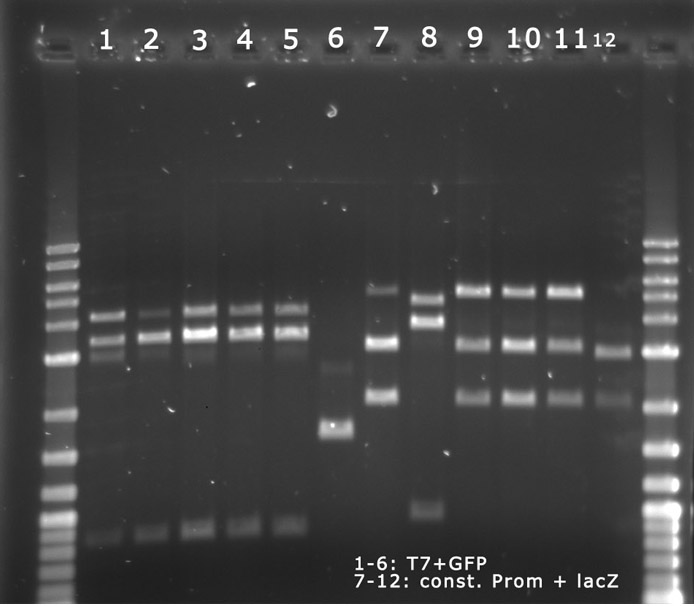
Analytical Digest of Minipreps
The above Minipreps from above were digested via Microwave digestion and applied onto a 0,8% Agarose gel.
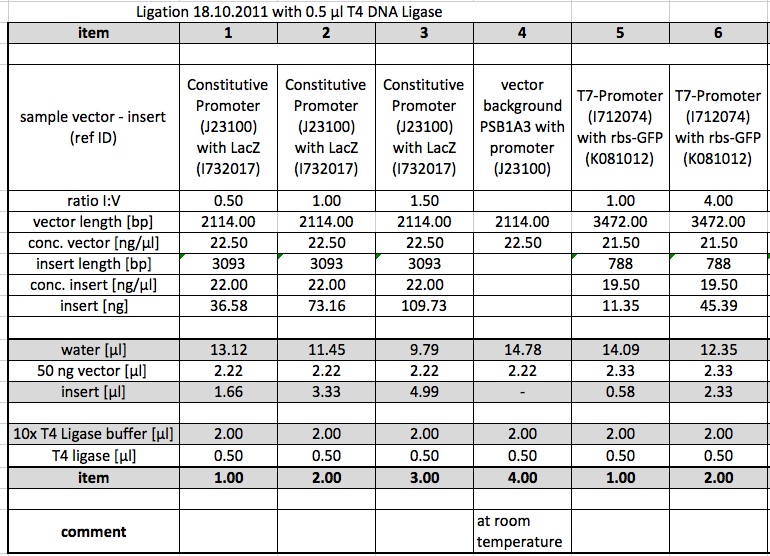
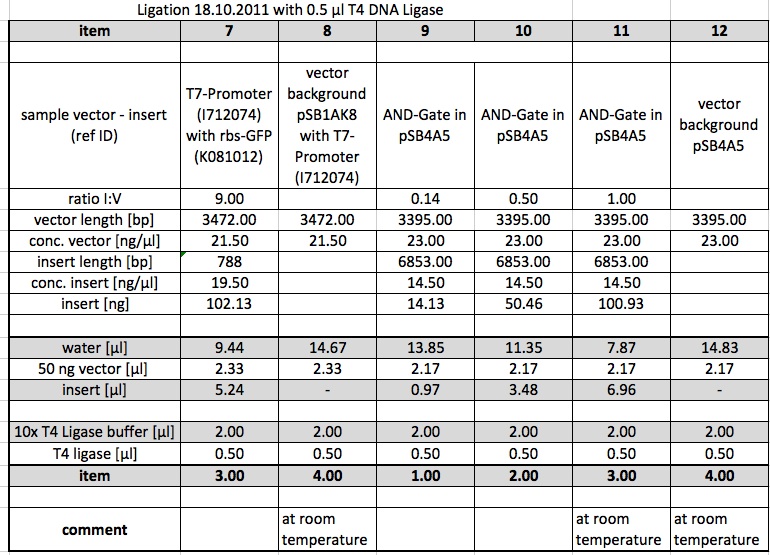
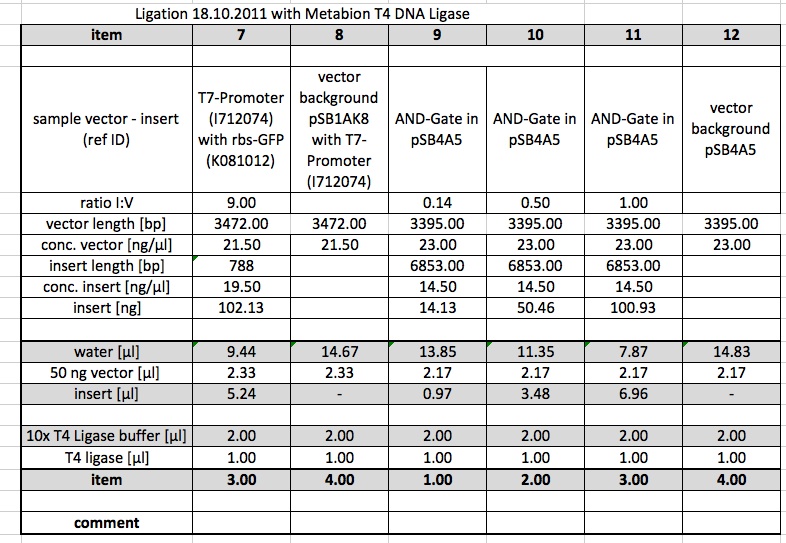
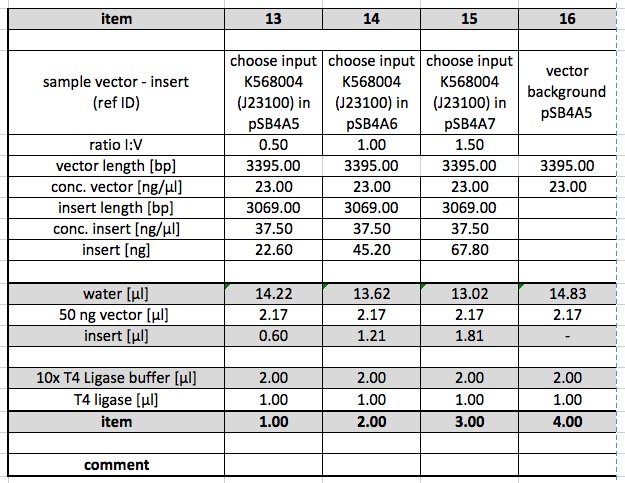
Ligations
The following Ligations were performed using the purified samples from the digestion from 15.10.11. The first ligations were made using 0.5 µl of the T4 DNA Ligase from NEB. Samples 4,8,11,12 were incubated at room temperature.
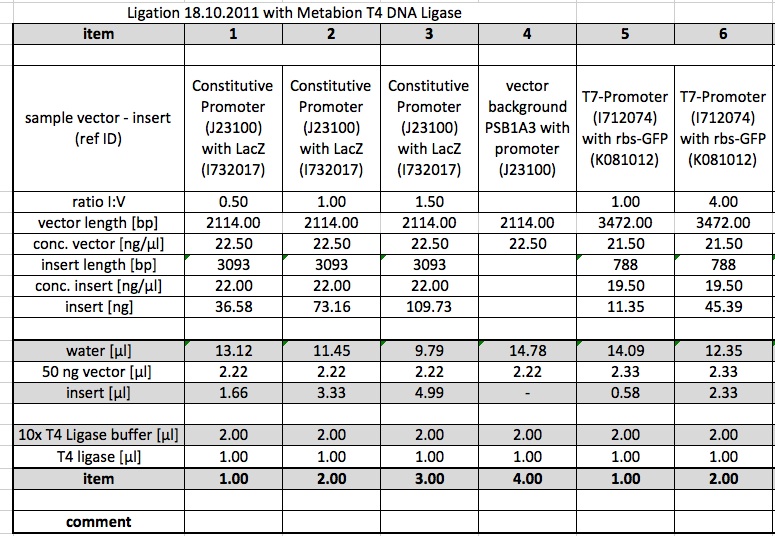
Ligations were repeated using 1 µl of the T4 DNA Ligase from metabion.
Restriction Digest
K568004 from Box T7, clone 6-2 (61.5 ng/µl) was digested using E,P. This is in order to ligate it into pSB4A5, which was already digested.
Ligation of K568004 with pSB4A5
Ligation of K568004 and pSB4A5 from the digestion of 15.10.11 were performed following this protocol:
Transformation
The part K568003 (T7-lacZ reporter plasmid; in pSB1AK8) was transformed into BL21(DE3) cells and plated on Amp plate over night at 37 °C. For the transformation, clone 2c 9.9.11 131 ng/µl was used.
Other Work
Media
Kan, Cam and Amp LB plates were made
S-gal plates with Amp and LB were made
LB liquid media was made
19-10-2011
People: Thorsten, Susan, Marta, Leah, Bea, Wolfgang
Other Work
Picking
-K568003 in BL21DE3: Plate from 18.10.11 Trafo T7-LacZ clone 2 in BL21 (amp)
-Red Light with GFP: Plate from 13.10.11 Ligation 15 (cam)
-Constitutive Promoter + GFP : Plate from 12.10.11 Trafo Ligation 10 in DH5alpha (amp)
-pBad/araC I0500 in DH5alpha
Digestion
Digestion of the plasmid preps from 18.10.11 with: 1 µl DNA
1 µl SpeI
1 µl Pst
7 µl water
for 1 h in 37°C
Analyt. gel run; 130 V, 1,5 h
Glycogn-ethanol-precipitation
Ligations from 18.10.11, but only from the samples where the Metabion T4 ligase was used
Ligation
The Ligation Number 4 (the control, which would have been just the plasmid) was not performed, because there wasn't enough DNA left.
Transformations
in JT2:
ID:10 Lig.3: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 205,0 ng/µl: of Mini-prep (Metabion) of the overnight cultures from the 17.10.11. --> Amp
RBS in pSB1A2 (for negativ control) --> Amp
in DH5alpha:
Ligations of 18.10.11 with following ID's: 9-12 (AND-Gate in low copy pSB4A5) and 13-16 (choose first input in pSB4A5) (1-8 was frozen @ -20°C) --> Amp
in BL21(DE3)
ID:10 Lig.3: const.prom+LacZ in DH5alpha Amp (Ligation 14.10.11) 205,0 ng/µl: of Mini-prep (Metabion) of the overnight cultures from the 17.10.11. --> Amp
20-10-2011
People: Susan, Leah, Bea, Wolfgang, Alex
Cloning
Mini-Prep
| Part | Clone | Concentration [ng/µl] |
|---|---|---|
| Lig. 10 from 15-10-2011 LB+Amp:const-prom+rbs GFP | 1 | 128 |
| 3 | 122 | |
| 4 | 136 | |
| Lig. 15 from 15.10.11: LB+Cm red light+rbs-GFP | 1 | 131 |
| 2 | 154 | |
| 3 | 107 | |
| 4 | 111 | |
| pBad Ara DH5alpha I0500 19-10 Kan | 1 | 135 |
| 2 | 152 |
Clone 2 of 19-10-2011 Lig. 10 in DH5alpha LB+Amp did not yield a culture of bacteria.
Digestion
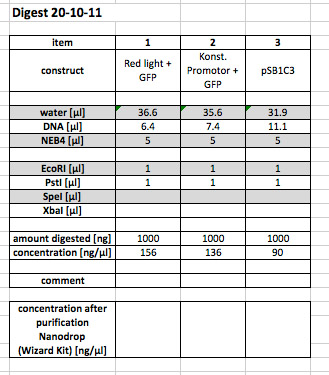
restriction digestion of Red light + GFP and const. promoter with E and P to subclone it in pSB1C3 (also cut with E and P)
prep. gel
purified dna is stored in the "igem box 1" in one corner
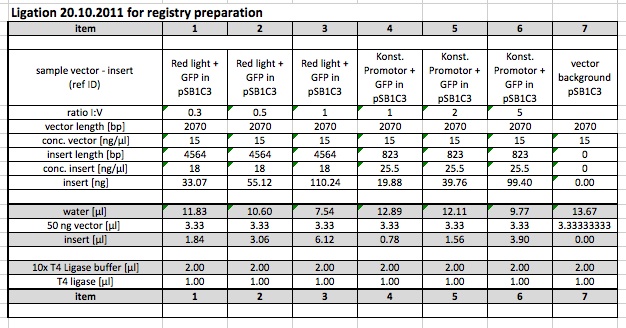
Ligation
Glycogen/Ethanol Precipitation and Transformation
Ligations from 19.10. were concentrated and purified via glycogen ethanol precipitation.
1-3 (T7-GFP) are transformed into DH5alpha
5-8 were transformed into JT2
plated on LB+Cm (1-3) LB+ Amp (5-8)
Other Work
Gylcerol Stocks
Glycerol stocks have been prepared from the following strains: Lig. 10 15.10.11: const-prom+rbs GFP in DH5alpha LB+Amp (clone 1); Lig. 15 15.10.11: red light+rbs-GFP LB+Cm (clone 1) in DH5alpha; 19.10.11 LB+Amp Trafo T7-lacZ in BL21 clone 2 K568003; pBad Ara DH5alpha I0500 19.10. Kan (clone 1).
Electrocompetent Cells
5 ml of over night culture TOP10f' cells were inoculated into 250 ml LB. However, we might not need this strain after all.
Preparations for lacZ - Reporter construct test
5ml LB+ Amp K568003 Bl21 DE3 (inoculation with o.n.culture from 19.10) 5ml Lb+ Amp LacZ without promoter in Bl21 DE3 (inoculation with glycerol stock) 5ml LB+ Kan T7 promoter in BL21 De3 (inoculation with glycerol stock)
Transformation
Red light+GFP and const. promoter + GFP (Minipreps 20.10.) 1ul each in JT2 cells
Picking
-const.prom + LacZ in BL21DE3, ID10 Lig.3 vom 19.10.11 (amp)
-Nur Vektor mit AB in JT2,RBS in PSB1A2 vom 19.10.11 (amp)
-AND-Gate in low copy Vektor (PSB4A5) JT2, Lig.9 vom 18.10.11 in DH5alpha (amp)
-AND-Gate in low copy Vektor (PSB4A5) JT2, Lig.10 vom 18.10.11 in DH5alpha (amp)
-AND-Gate in low copy Vektor (PSB4A5) JT2, Lig.11 vom 18.10.11 in DH5alpha (amp)
-K568004, Lig.14 vom 18.10.11 in DH5alpha (amp)
-K568004, Lig.15 vom 18.10.11 in DH5alpha (amp)
-const.prom + LacZ in JT2, ID10 Lig.3 vom 19.10.11 (amp)
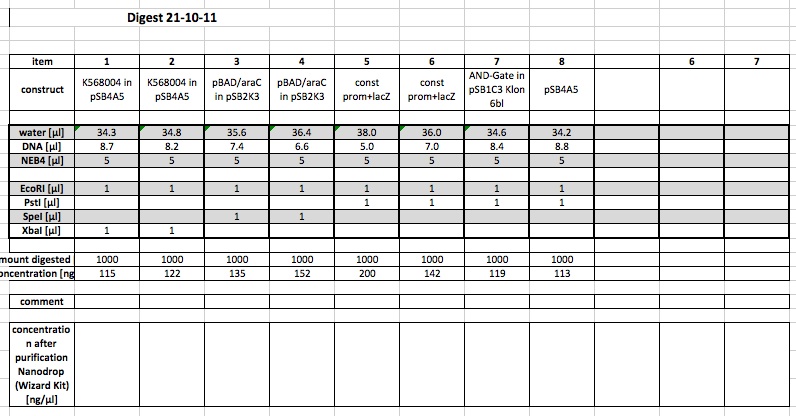
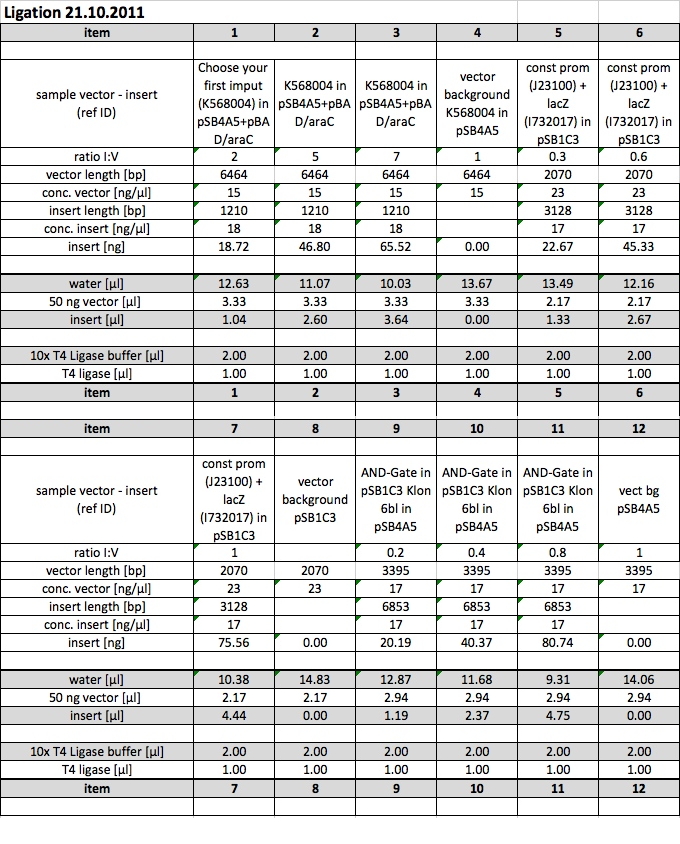
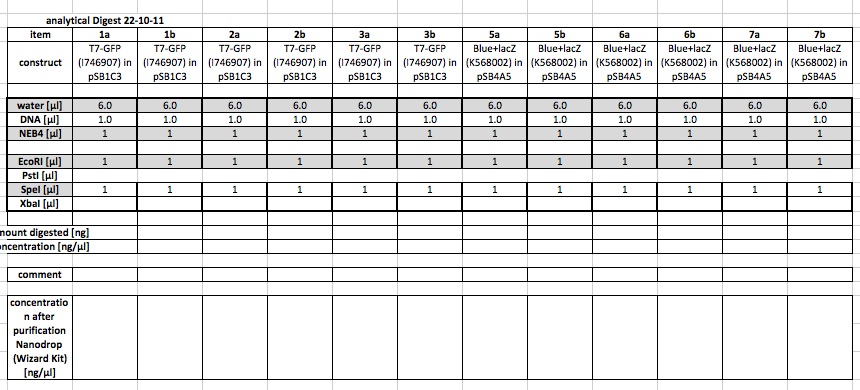
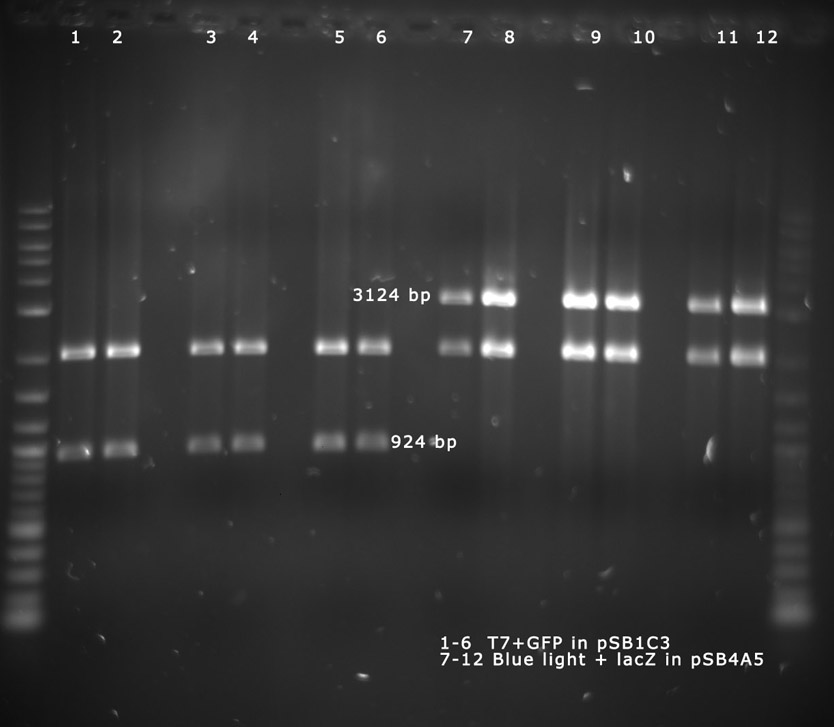
21-10-11
People: Bea, Leah, Wolfgang, Alex, Marta, Thorsten
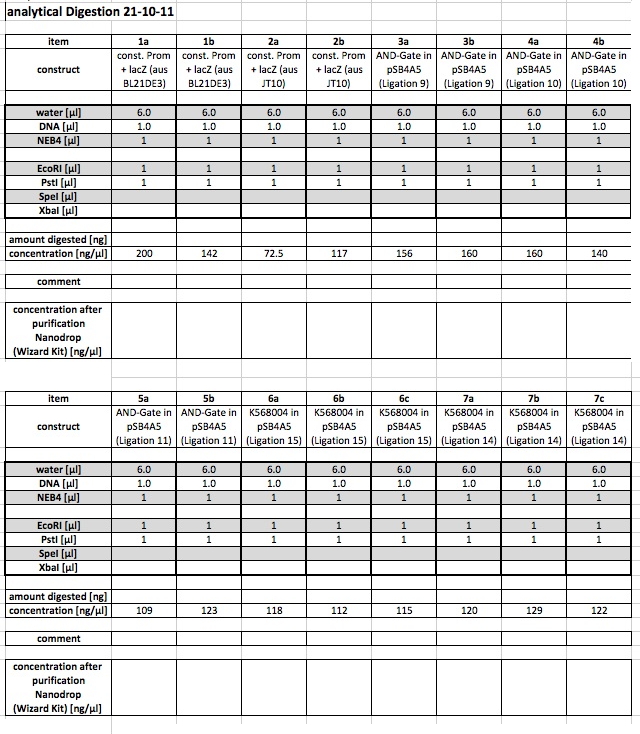
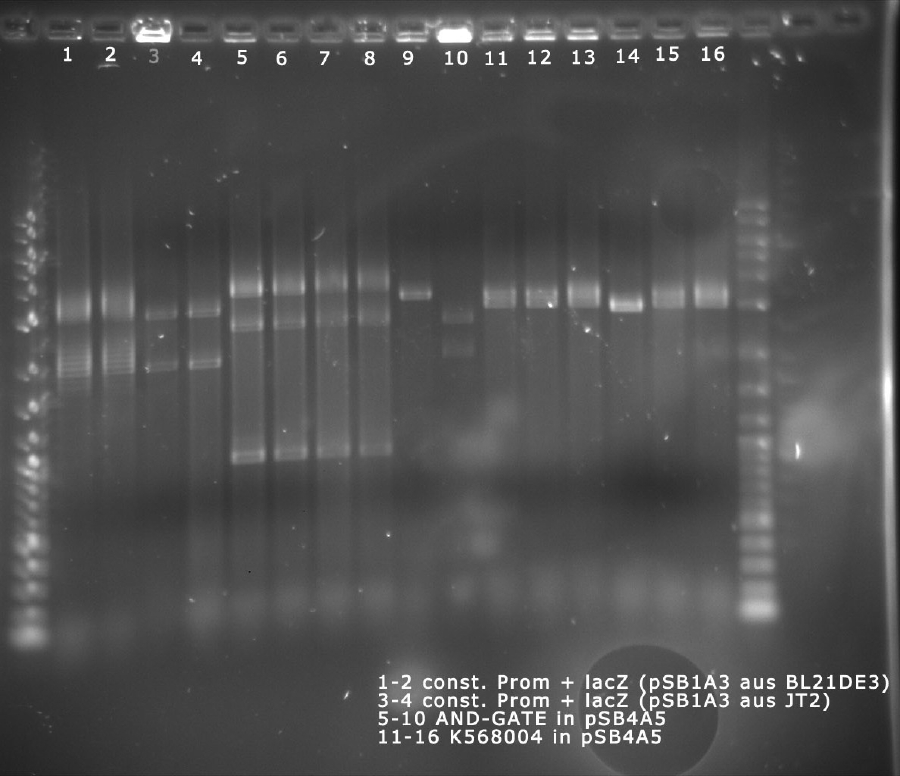
analytical digest
The following ligations were digested analytically:
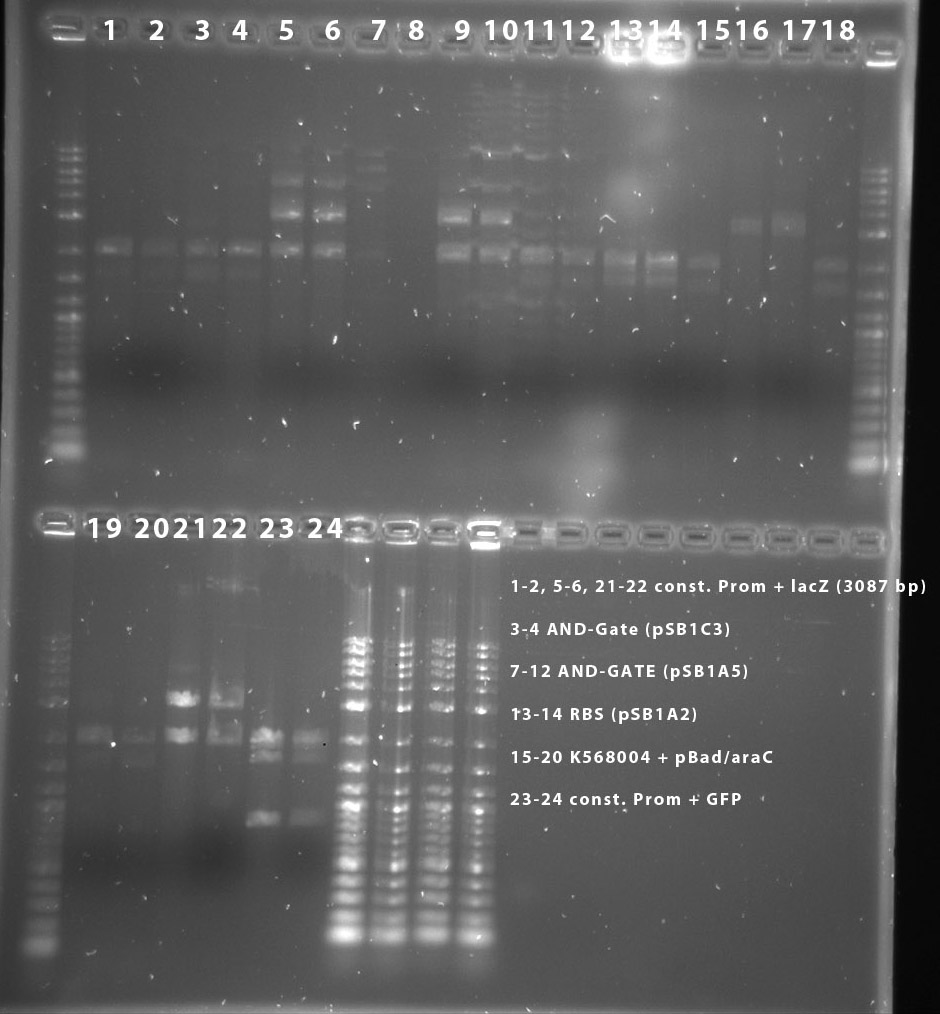
The digest shows that the ligation of the constituive promotor and lacZ worked.
The digest shows that the ligation of the AND-Gate into pSB4A5 seems to have failed. This has to be repeated.
The following samples were digested preparative:
using 1a and 1 b and 6c and 7c for further work:
Ligation:
 "
"