Team:USTC-China/Project/module
From 2011.igem.org
(Difference between revisions)
(→Riboswitch) |
|||
| Line 11: | Line 11: | ||
==Riboswitch== | ==Riboswitch== | ||
[[File:Riboswitch.jpg|center]]<br/> | [[File:Riboswitch.jpg|center]]<br/> | ||
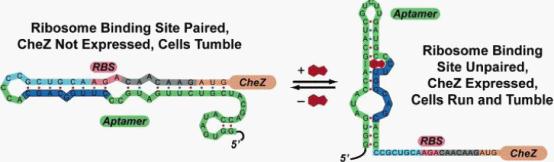
| - | </font><font size=" | + | </font><font size="2">Figure 1: Model for how the theophylline-sensitive synthetic riboswitch controls the translation of the CheZ protein. In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811)</p></font><font size="3"> |
<html><a name="Toggle-switch" id=""></a></html> | <html><a name="Toggle-switch" id=""></a></html> | ||
Revision as of 20:33, 5 October 2011

Contents |
Modules
1.Riboswitch
2.Toggle-switch
3.Quorum Sensing
4.Artificial Innate Immunity System
Riboswitch
Figure 1: Model for how the theophylline-sensitive synthetic riboswitch controls the translation of the CheZ protein. In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811)</p>
Toggle-switch
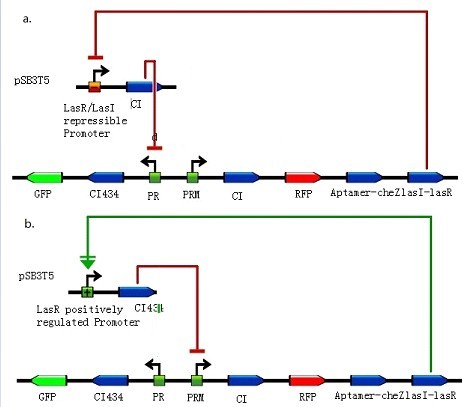
Figure 2: Toggle switches incorporate three mechanisms: positive feedback, double-negative feedback, and the repressor binding cooperatively and two different kinds of bacteria differentiate out of homogeneous conditions. (Images from: Chunbo Lou, Xili Liu, Ming Ni.etc (2010) Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Molecular Systems Biology, 6:350) Figure 3: Constructions to modulate the ratio of toggle switches. The above bar represents pSB3T5, a low to medium copy BioBrick standard vector, where LuxpR (BBa_R0062) controls the expression of cI repressor which would influence the ratio of toggle switch. (Images by USTC-IGEM with TinkerCell)
 "
"