Team:BU Wellesley Software/Notebook/KyleNotebook
From 2011.igem.org
| Line 77: | Line 77: | ||
6/22: Today I set up a gel electrophoresis of restriction digest samples. The samples were E0430 (YFP composite), E0240 (GFP composite), R2000 (constitutive promoter), and B0015 (terminator). Each sample took two wells and were followed by their uncut plasmids. | 6/22: Today I set up a gel electrophoresis of restriction digest samples. The samples were E0430 (YFP composite), E0240 (GFP composite), R2000 (constitutive promoter), and B0015 (terminator). Each sample took two wells and were followed by their uncut plasmids. | ||
| + | |||
{| | {| | ||
| - | |[[Image:6_22_11_restrictiondigest1.jpg | | + | |[[Image:6_22_11_restrictiondigest1.jpg | 200px]] |
|} | |} | ||
| + | |||
We also had our Wet lab meeting today in which we discussed our division of work: a team to work on combining the composite parts with promoters and another team to start making constructs with our fluorescent protein genes (BFP, RFP, EYFP, ECFP). | We also had our Wet lab meeting today in which we discussed our division of work: a team to work on combining the composite parts with promoters and another team to start making constructs with our fluorescent protein genes (BFP, RFP, EYFP, ECFP). | ||
Revision as of 16:59, 1 July 2011
Contents |
Current To Do List
This is my online notebook. Check here for weekly updates on what I am doing in the lab and how things are going.
Weekly Journal
6/6-6/10
As the first week of actual iGEM work, there is some initial prep work that needs to be done. Before we can start focusing on the main project involving the development of DNA plasmids and invertases to function within the cell, we must confirm our protocols and create working plasmids with fluorescent protein genes. Also, we want to see which promoters and ribosomal binding sites (RBS) work the best together.
6/6: Monday was our last day of "boot camp". With the comp. team and Wellesley students, we discussed basic synthetic biology and the different procedures/protocols that we would be using in the lab. We also reviewed a paper from Science magazine, Synthetic Gene Networks that Count by Friedland, et al. A tour of the lab concluded the day.
6/7: In the first day in the lab, I did a liquid culture plasmid prep of plated transformations that our Post-Doc mentor Traci had already done. These would go into the incubator and be ready for mini-prep and plasmid isolation tomorrow. As a wet lab team, we browsed the Registry of Standard Biological Parts and chose the parts that we would work with over the next couple of weeks. In all, we chose 3 constitutive promoters, 2 inducible promoters, and 3 RBS, each of varying strength. We subsequently went through the transformation protocol and inserted the plasmids into competent cells. These would incubate overnight.
6/8: Today I did mini-prep and plasmid isolation on the liquid cultures from the day before. The DNA quantification of these samples proved to be poor, with only a few having viable concentrations of DNA for further use. Additionally, we ran a gel electrophoresis of the plasmid samples. The gel showed no bands of DNA, thus further confirming the absence of viable plasmid DNA from the samples. This result is causing some consternation as the exact reason for the problem is still unknown to us. The transformation plates of our selected parts were successful. The rest of the day was spent doing liquid culture plasmid preps of these plates that we had transformed on 6/7.
6/9: Today we did the mini-prep and plasmid isolation of our transformation of selected parts(3 const. promoters, 2 ind. promoters, and 3 RBS). The DNA quantification of these samples were poor (like our samples the day before) and a gel electrophoresis of the plasmids gave indication of little to no plasmid DNA. Only one part (UV light promoter) had a viable DNA concentration. One thing we noticed is that the samples did not grow well in the liquid cultures and there was almost a stringy characteristic to the cells in the tube. For further investigate, we went to the Biology labs and performed Gramm Staining on 6 of the samples. What we saw under the microscope was some cells that resembled E. Coli along with bacteria in a long string/beadlike form. Our conclusion was that the samples were contaminated and that we needed to be more conscientious about lab cleanliness.
6/10: There was not much work in the lab today. I autoclaved some pipet tips that we would use. Instead, the day was spent organizing how we would put together our different BioBrick parts into a full working construct plasmid. This included how to take out the promoters in the original RBS plasmids, as well as which restriction enzymes to use. We planned out the next week and assigned tasks to everyone.
6/12-6/18
This week, as a team, we will combine the GFP gene and terminator into one plasmid. Also we will continue to troubleshoot and figure out what went wrong with our transformations from Week 1. If we need to, we will set up new transformations of our selected BioBrick parts.
6/12: Came in on a Sunday to do liquid cultures plasmid prep of our original plated transformations of our selected BioBricks (5 promoters and 3 RBS). Our theory is that contamination occurred when we made our liquid cultures the last time. So we were careful in our aspectic technique and made two liquid cultures: one with LB, and one with LB + ampicillin.
6/13: This morning I checked on the liquid plasmid preps from 6/12 and found that only two of the LB+Amp cultures grew while all of the LB cultures grew. This meant that our transformation plates from the previous week were no longer selective to our Amp resistant transformed E. coli and were no usable anymore. I did minipreps on the two cultures that did grow (an RBS and UV light inducible promoter), but they had poor DNA concentration like the week before. So we did totally new transformations of the RBS and promoters of which we had DNA left in the 2010 iGEM plates.
6/14: The transformations from yesterday grew overnight and I checked them this morning. The negative control plate we did (T10 E. coli cells only) grew a lot of colonies, thus effectively pinpointing the source of our problems the past week. The agar plates which had ampicillin on them were no longer effective in killing all other bacteria besides what we had transformed which carried resistance to the antibiotic. Hence why we had such contamination and poor isolation of plasmid DNA. The other plates looks abnormal for E. coli colonies and they were thrown out. The Kanamycin resistant plates worked fine and had no contamination. We proceeded with plasmid preps for the two samples that were grown on Kanamycin plates, the GFP+Terminator (BBa_J52028+ BBa_B0015) composite part and the constitutive promoter BBa_I14033.
|
Contaminated E. coli plate
Another thing that came on 6/14 was the QIAcube, a robotic machine that automates all QIAgen protocols. We are demoing the machine to see if it is worth purchasing. We will use the machine to do our mini prep and gel extraction protocols. See the QIAgen website for a more detailed description and video: http://www.qiagen.com/products/automation/qiacube.aspx

|
QIAcube
6/15: Today I did the plasmid preps from the GFP+Term composite part and the K-resistant promoter from yesterday. DNA quantification was overall decent, much better than the previous week. Also, I made new agar plates with ampicillin. Luckily the 2011 iGEM plates with the DNA arrived and we transformed our promoters and RBS parts again using the new materials. We transformed the same 5 promoters, 3 RBS, and the RFP gene. Today was also our lab meeting which was run by one of our graduate student mentors, Suma. We discussed our recent problems and the steps we've been taking to fix them. As a team, we decided on transforming another promoter (BBa_R2000) and GFP composite part (BBa_E0240) so that we could use Kanamycin plates and quickly develop a working GFP construct plasmid.
6/16: This morning the transformation plates of the 5 promoters, 3 RBS, and RFP looked good. Most had a few colonies and some had a lot. Only the inducible UV light promoter (BBa_I765001) had no colonies (as well as the negative controls). Liquid culture plasmid preps were done on the plates that grew. In attempt to make more GFP+Term composite parts which we will need to combine with the various RBS and promoter parts, I did a restriction digest, gel electrophoresis, and gel extraction. It will be ligated later today and then transformed along with our new parts tomorrow.
6/20-6/24
6/20: This morning I set up the QIAcube to do minipreps of the liquid plasmid cultures of our transformed reporters (RFP BBa_E1010, EYFP BBa_E0032, ECFP BBa_0020, BFP BBa_K156010) and composite parts (GFPc BBa_E0240 and YFPc BBa_E0430) as well as a promoter (BBa_R2000) chosen for the composite parts. The DNA quantification was good, with most numbers around 30-50 ng/ul. In anticipation of needing more promoter and RBS plasmid DNA for our constructs, I used the transformation plates from 6/15/11 to make liquid plasmid preps. The rest of the afternoon was spent planning for our BU iGEM meeting and what we would present as a wet lab team.
6/21: This morning I prepared minipreps of the promoter and RBS liquid cultures from yesterday for the QIAcube to run. Today was our BU iGEM wet and dry lab meeting. As our first one, it involved an overall presentation on what has been happening in each of the teams. As a wet lab team, our presentation was more group as a lot of what we had done, everyone had been apart of. We discussed our initial contamination problems, the parts that we have chosen, and our current success. Also we developed a timeline of events:
June: - 4 “devices” PCR TB Genes (made-tested-verif/Clotho)
- Teams (assigned, robot learning, RD/ligation)
- Picked “n” invertase (have) - PCR TB Genes
July: - Get invertase ready - Make 4 devices with invertase... Test - Robot puts together devices (RD/ligation 2 parts) - Manual Reconfig - Robot puts devices together - Robot Transformation
6/22: Today I set up a gel electrophoresis of restriction digest samples. The samples were E0430 (YFP composite), E0240 (GFP composite), R2000 (constitutive promoter), and B0015 (terminator). Each sample took two wells and were followed by their uncut plasmids.
| 200px |
We also had our Wet lab meeting today in which we discussed our division of work: a team to work on combining the composite parts with promoters and another team to start making constructs with our fluorescent protein genes (BFP, RFP, EYFP, ECFP).
6/23: This morning I did restriction digest of our GFP+term composite part, J61101 (RBS), J61100 (RBS), J61127 (RBS), and K156010 (BFP). The RBS's I cut with Spe1 and Pst1 while the BFP I cut with EcoR1 and Spe1. The GFP+term was cut with Xba1 and Pst1. A gel electrophoresis was run. The GFP+term still only showed one band, the same result we had two days earlier. Thus we decided that the restriction digest was not the conflict here and rather the ligation did not work properly. With the BFP and RBS samples, the separation looked good and the samples ( lower bands for BFP corresponding to the smaller insert and higher bands for RBS corresponding to the large backbone) were cut out and purified through gel extraction quickprep on the QIAcube.
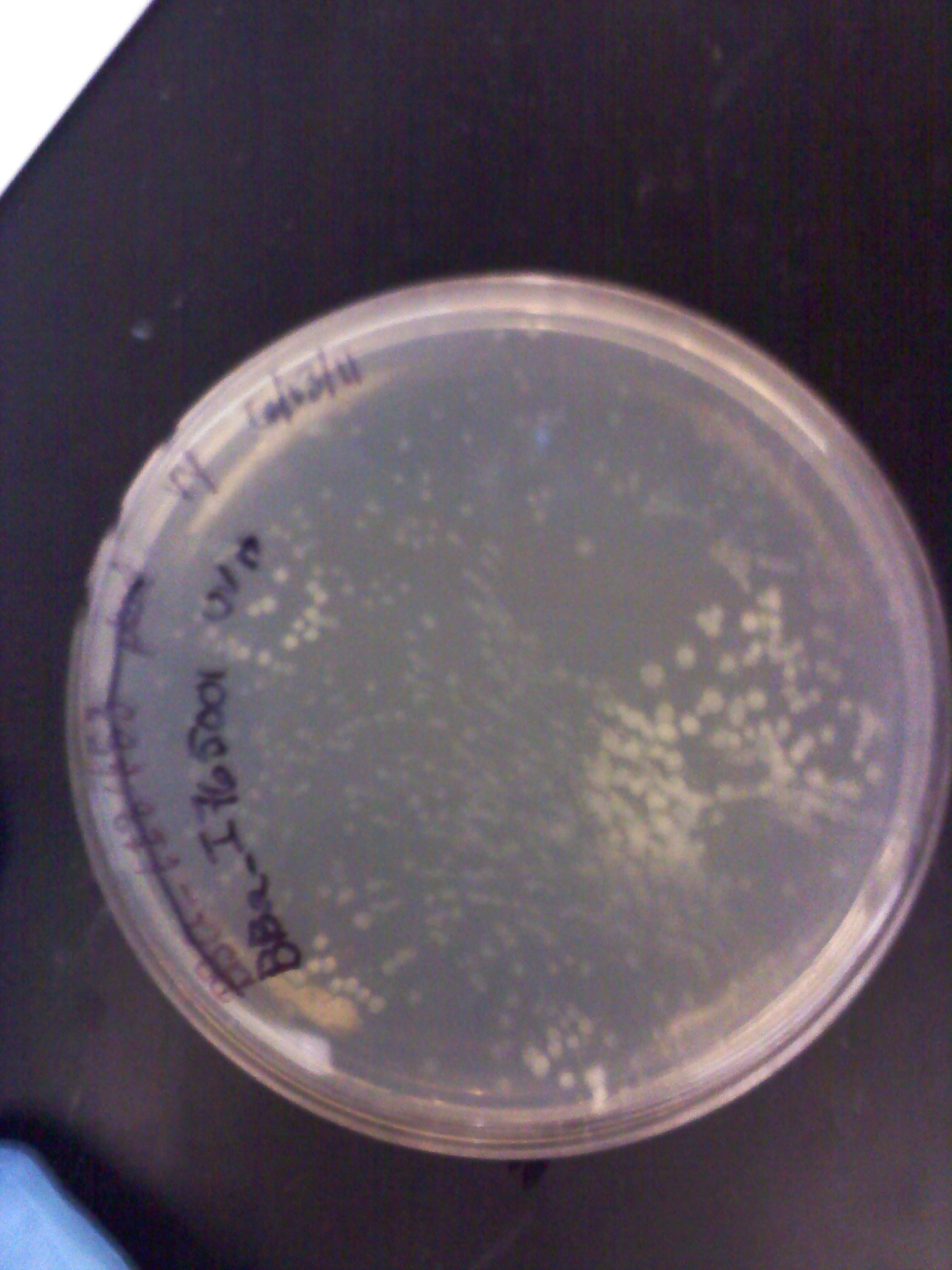
6/24: Today I started the day with a restriction digest of RFP, ECFP, EYFP, and RBS (J61101 and J61100). The samples were loaded into a gel respectively, each followed by their uncut plasmids.
| 300px |
For some of the uncut plasmids, not enough sample was put into the well, so they are hard to see. For all the fluorescent proteins, the enzymes EcoR1 and Spe1 were used and thus the lower bands on the gel were removed. The RBS's were cut with Spe1 and Pst1 and the higher bands were removed.
Because we were running out of RBS plasmids and the promoter I14033, I used the frozen glycerol stock of our transformed cells to plate and grow overnight for plasmid preps over the weekend.
 "
"