Team:Utah State/Project
From 2011.igem.org
(→Results) |
Cwbarentine (Talk | contribs) |
||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/7/7c/Headers3.png" width="739" height="29" border="0" usemap="#Map"> | ||
| + | <map name="Map"> | ||
| + | <area shape="rect" coords="-1,-2,70,27" href="https://2011.igem.org/Team:Utah_State#CyanoBricks"> | ||
| + | <area shape="rect" coords="74,-9,132,28" href="https://2011.igem.org/Team:Utah_State/Team"> | ||
| + | <area shape="rect" coords="142,-4,269,27" href="https://igem.org/Team.cgi?id=654"> | ||
| + | <area shape="rect" coords="284,-1,363,31" href="https://2011.igem.org/Team:Utah_State/Project"> | ||
| + | <area shape="rect" coords="379,-3,447,30" href="https://2011.igem.org/Team:Utah_State/Safety"> | ||
| + | <area shape="rect" coords="464,-15,595,28" href="https://2011.igem.org/Team:Utah_State/Attributions"> | ||
| + | <area shape="rect" coords="611,-1,716,27" href="https://2011.igem.org/Team:Utah_State/Parts"> | ||
| + | </map> | ||
| + | </html> | ||
<html> | <html> | ||
<style type="text/css"> | <style type="text/css"> | ||
| Line 28: | Line 40: | ||
</style> | </style> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</html> | </html> | ||
| - | |||
== '''Overall project''' == | == '''Overall project''' == | ||
| Line 126: | Line 119: | ||
<html> | <html> | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/7/7c/Headers3.png" width="739" height="29" border="0" usemap="#Map"> |
| - | < | + | <map name="Map"> |
| - | < | + | <area shape="rect" coords="-1,-2,70,27" href="https://2011.igem.org/Team:Utah_State#CyanoBricks"> |
| - | + | <area shape="rect" coords="74,-9,132,28" href="https://2011.igem.org/Team:Utah_State/Team"> | |
| - | < | + | <area shape="rect" coords="142,-4,269,27" href="https://igem.org/Team.cgi?id=654"> |
| - | < | + | <area shape="rect" coords="284,-1,363,31" href="https://2011.igem.org/Team:Utah_State/Project"> |
| - | + | <area shape="rect" coords="379,-3,447,30" href="https://2011.igem.org/Team:Utah_State/Safety"> | |
| - | < | + | <area shape="rect" coords="464,-15,595,28" href="https://2011.igem.org/Team:Utah_State/Attributions"> |
| - | + | <area shape="rect" coords="611,-1,716,27" href="https://2011.igem.org/Team:Utah_State/Parts"> | |
| - | + | </map> | |
| - | < | + | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | + | ||
| - | + | ||
| - | </ | + | |
</html> | </html> | ||
Revision as of 23:43, 28 September 2011
![]()
Contents |
Overall project
Abstract:
The field of Synthetic Biology is continuously increasing our understanding of the complexity of all living systems, one gene at a time. Our project focuses on producing high value bioproducts in the form of fatty alcohols, wax esters, and alkanes/alkenes using Synechocystis sp PCC 6803. In addition, our team is investigating the strength of 23 promoters by measuring a dual luciferase expression construct.
Project Description:
Building upon the CyanoBrick toolkit developed by the 2010 Utah State iGEM team, our project will focus on producing valuable bioproducts using Synechocystis sp. PCC 6803. Our project will attempt to produce 3 different bioproducts: fatty alcohols, wax esters, and alkanes/alkenes. Fatty alcohols and wax esters are used to produce cosmetics, lubricants, and various pharmaceuticals. Fatty alcohols with a 20-carbon chain are used in the cold sore medication Abreva[reference needed], and long chain fatty alcohols have been linked to improved heart health [reference needed]. Alkanes and alkenes are utilized as hydrocarbon fuels. The production of these bioproducts in cyanobacteria will greatly reduce their cost and increase their availability.
In addition to producing bioproducts, our project will also provide a more detailed characterization of the promoters and ribosome binding sites (RBS) produced by the 2010 Utah State iGEM Team. Utilizing a dual luciferase expression measurement construct, and reference promoters from E. coli and Synechocystis, our team will more precisely characterize the expression levels of 23 promoters under standard growth conditions, compared with the GFP-based measurements made in 2010. We will also produce useful intermediate parts, which are currently not available through the registry, allowing the dual luciferase expression measurement system to be easily adapted to new organisms and new reference promoters.
Justification:
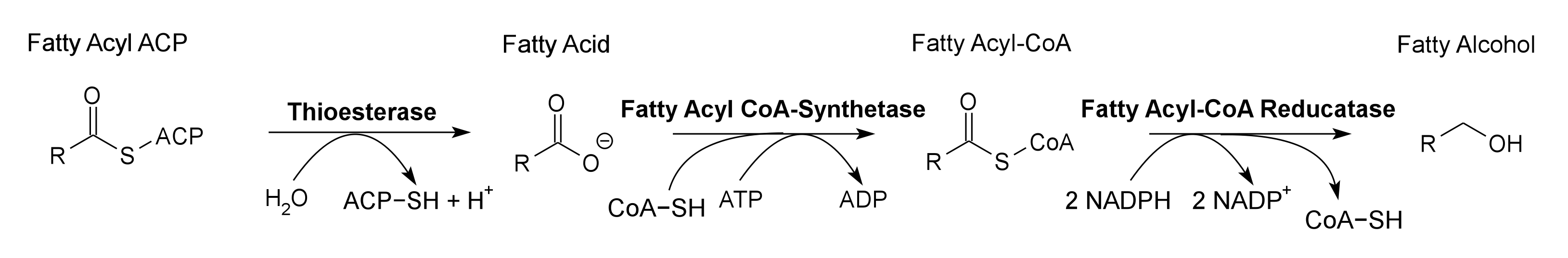
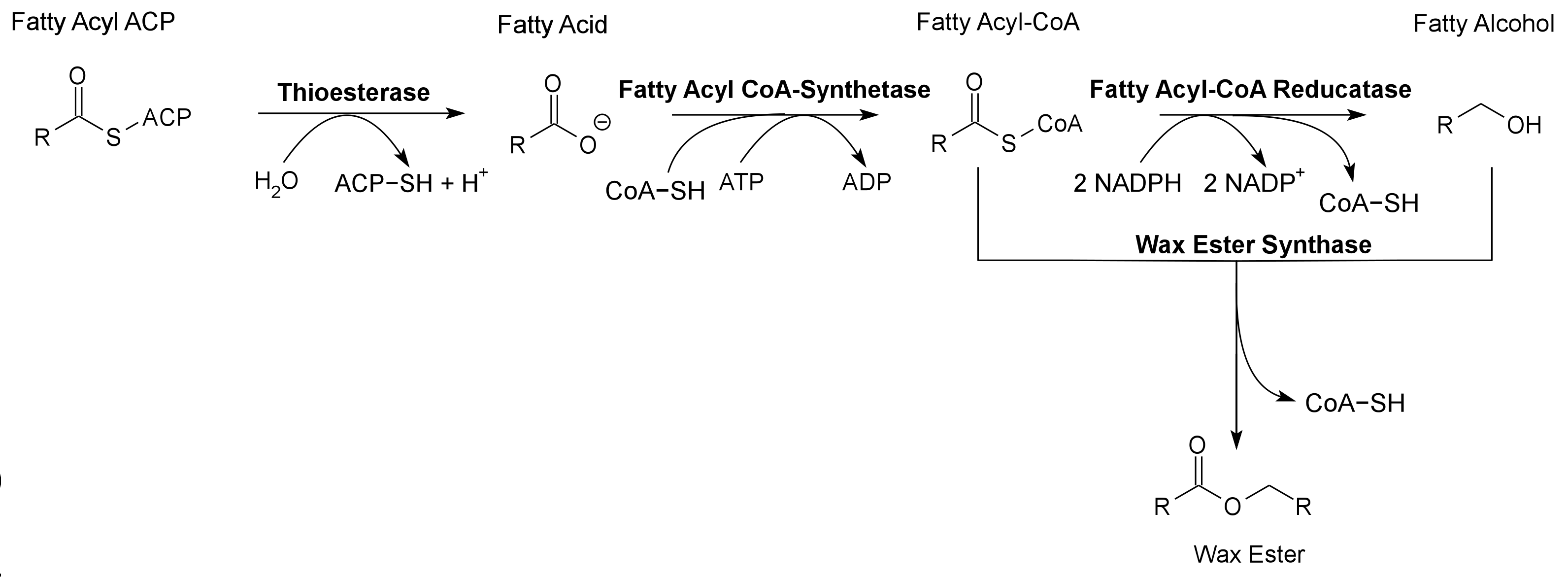
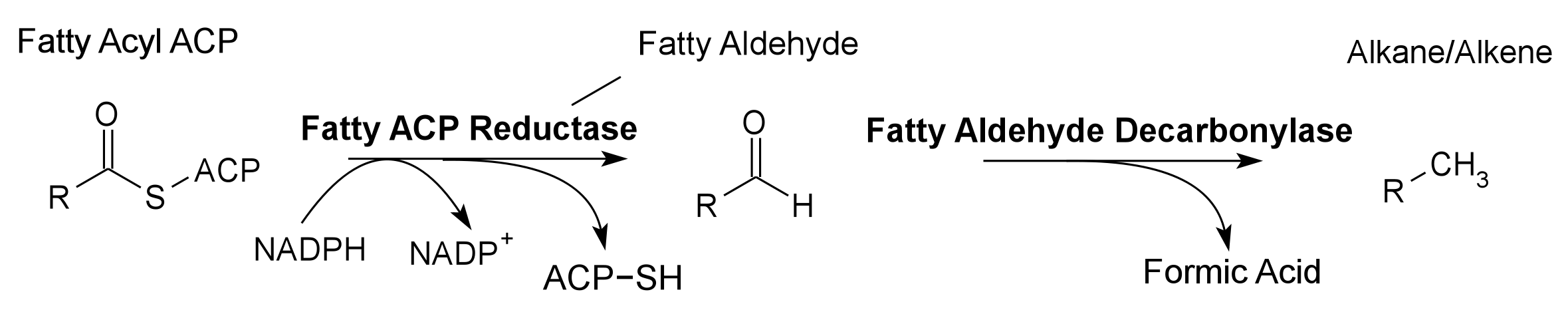
Global energy requirements are heavily dependent on fossil fuels such as oil, coal, and natural gas. Efforts to lower our dependence on foreign oil, along with the anticipation of fossil fuels being exhausted in the future, novel strategies need to be discovered for alternative energy generation. Renewable hydrocarbons (Schirmer et al., 2010), fatty alcohols, and wax esters (Keasling et al., 2010) may provide “drop in” compatible fuels and chemicals. The production of alkanes has been reported in a wide variety of microorganisms, particularly photoautotrophic cyanobacteria (Schirmer et al., 2010). Our group is focused on genetically engineering the necessary pathways needed to produce fatty alcohols (Figure 1), wax esters (Figure 2), and alkanes/alkenes (Figure 3) using Synechocystis sp PCC 6803 as the model organism. In addition, we are investigating the strength of 23 promoters by measuring a dual luciferase expression construct.
Synechocystis sp PCC 6803:
Synechocystis sp. PCC 6803 (hereafter referred to as PCC 6803) is a cyanobacterium, a photosynthetic bacterium, and one of the major model organisms in microbiology. The photosynthetic pathway of PCC 6803 has been extensively studied and its genome has been completely sequenced. It is capable of acting as both an autotroph, obtaining its energy from light and its carbon from the atmosphere, and as a heterotroph, using glycolysis of sugars as both an energy source and a carbon source. It also possesses the ability to uptake DNA present in its environment through natural transformation, making it a useful species for genetic manipulation (Zhang et al., 2007). PCC 6803 possesses many industrial applications including lipid recovery in biofuel production, PHB collection, and as a toxin biosensor (Rouillon et al., 1999; Wu et al., 2001). Heptadecane, an alkane hydrocarbon, production has been observed within PCC 6803 (Schirmer et al., 2010).
Project Details
Dual Luciferase
The dual luciferase system integrates two efficient reporter genes to evaluate regulatory gene expression. This system incorporates the firefly (Photinus pyralis) and the Renilla (Renilla reniformis) luciferases, which have different biochemical requirements for luminescence (Sherf et al., 1996). Dual reporter systems are commonly used to improve the accuracy of reporter assays when analyzing regulatory gene expression, particularly in mammalian cells. The luciferase assay is very sensitive assay that is performed by sequentially measuring both luciferase activities of the same sample (McNabb et al., 2005).
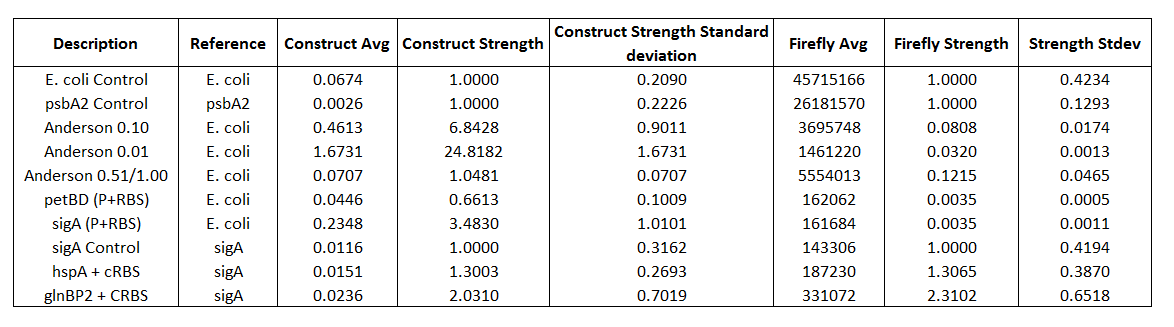
Promoters Tested
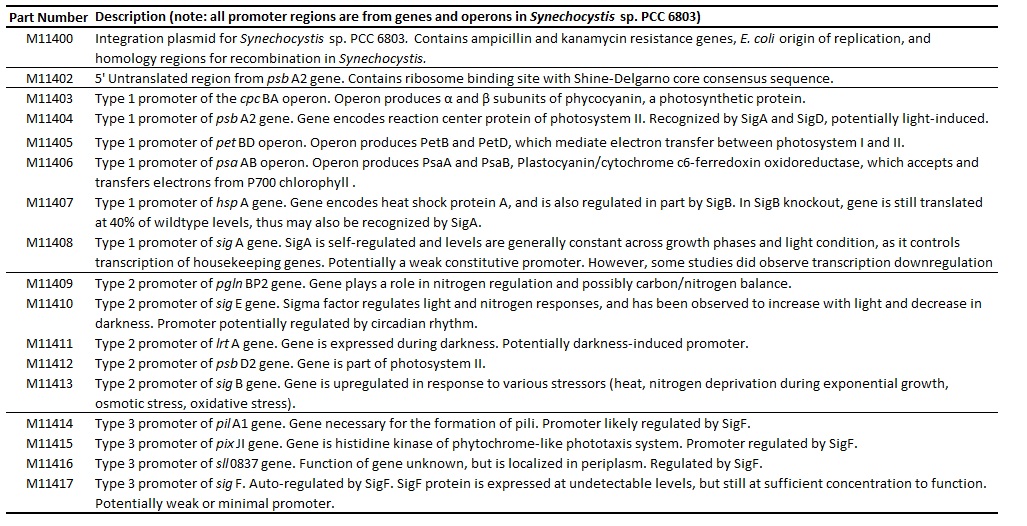
Table 1. Comprehensive list of all promoters tested.
Dual luciferase construct
Construct 1.
Bioproduct Pathways
Pathway 1 - Fatty Alcohol
Pathway 2 - Wax Ester
Pathway 3 - Alkane Alkene
Experiments
Results
Table 2. Construct Strength and Firefly Strength
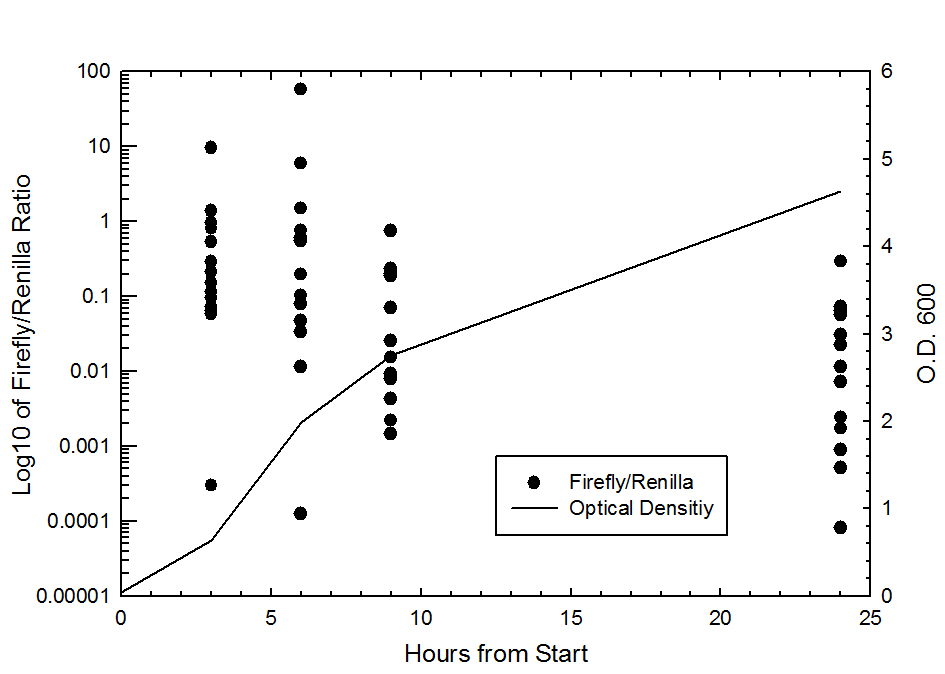
Graph of Firefly/Renila ratio vs. Time (hr)
References
1. Keasling, J.D., Steen, E.J., Kang, Y.S., Bokinsky, G., Hu, Z.H., Schirmer, A., McClure, A., del Cardayre, S.B., (2010) Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature 463, 559-U182.
2. McNabb, D.S., Reed, R., Marciniak, R.A., (2005) Dual luciferase assay system for rapid assessment of gene expression in Saccharomyces cerevisiae. Eukaryot Cell 4, 1539-1549.
3. Rouillon, R., Avramescu, A., Carpentier, R., (1999) Potential for use of a cyanobacterium Synechocystis sp immobilized in poly(vinylalcohol): Application to the detection of pollutants. Biotechnol Tech 13, 559-562.
4. Schirmer, A., Rude, M.A., Li, X.Z., Popova, E., del Cardayre, S.B., (2010) Microbial Biosynthesis of Alkanes. Science 329, 559-562.
5. Sherf, B. A., S. L. Navarro, R.R. Hannah, and K. V. Wood. 1996. Dualluciferase reporter assay: an advanced co-reporter technology integrating firefly and Renilla luciferase assays. Promega Notes 57:2–8. [Online.] http://www.promega.com/pnotes/57/5573a/5573a.html.
6. Wu, G.F., Wu, Q.Y., Shen, Z.Y., (2001) Accumulation of poly-beta-hydroxybutyrate in cyanobacterium Synechocystis sp PCC6803. Bioresour Technol 76, 85-90.
7. Zhang, X.C., Zang, X.N., Liu, B., Liu, S.M., Arunakumara, K.K.I.U., (2007) Optimum conditions for transformation of Synechocystis sp PCC 6803. J Microbiol 45, 241-245.
![]()
 "
"