Team:MIT/Notebook/
From 2011.igem.org
(Difference between revisions)
(→Overview) |
|||
| Line 921: | Line 921: | ||
<!----------------------------------------------------------------------------------------------------------------> | <!----------------------------------------------------------------------------------------------------------------> | ||
</!> | </!> | ||
| + | <h1> Week 11 </h1> | ||
| + | h3. &nbsp;August 21 | ||
| + | |||
| + | |||
| + | |||
| + | h4. Kenneth - Updates on Experiments: | ||
| + | |||
| + | |||
| + | |||
| + | Latest iteration of base FACS code: (incorporates Deepak's histogram update, removes the mean line on the histograms. Moves the quadrant lines to 200 for everything, since I've been seeing 200 as pretty much the max for the Blank samples. Can be easily changed. | ||
| + | |||
| + | |||
| + | |||
| + | [^auto_facsold.m]&nbsp;(the name is deceptive) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Results are in for dox ladder of Elowitz CMV-TO CHO cells [iGEM2011:Elowitz CHO Dox ladder], free delta induction of Elowitz receiver CHO cells.&nbsp;[iGEM2011:Free Delta Induction of Elowitz Receiver CHO] | ||
| + | |||
| + | |||
| + | |||
| + | FACS data is in for TRE:delta mcherry [TRE_Delta-mCherry dox ladder part 2|iGEM2011:TRE_Delta-mCherry dox ladder part 2]&nbsp;and TRE:mKate experiments &nbsp;[TRE_mKate dox ladder part 2|iGEM2011:TRE_mKate dox ladder part 2], however, due to loss of blue channel, cannot gate. Instead, using the top X% percent method to process data. The MATLAB code takes in the data and only keeps the top X% of data. In this case I used 50%. The program takes in filename, color to use as cutoff (in this case 'r'), and X for top X% (in this case 50).[^auto_facstop.m] | ||
| + | |||
| + | |||
| + | |||
| + | Co-Culture of Elowitz CHO cells: taken pictures also at 48 hours. Will bring to Weiss lab to process images. Redoing the experiment again, this time with 80:20 ratio as well. Will take pictures and FACS on Tuesday | ||
| + | |||
| + | |||
| + | |||
| + | CO-Culture of HEK and CHO cells:&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | \-Delta Heks and receiver CHO's: co-cultured Friday. added dox saturday. Will FACS Monday | ||
| + | |||
| + | |||
| + | |||
| + | \-Delta CHO's and receiver Hek's: same as above | ||
| + | |||
| + | |||
| + | |||
| + | \-TRE:delta Hek senders and CHO receivers: didn't have the resources to do just yet. Set up for experiment on Tuesday. | ||
| + | |||
| + | |||
| + | |||
| + | h5. Tyler | ||
| + | |||
| + | |||
| + | |||
| + | I just put up&nbsp;[iGEM2011:CHO Experiments]. I will try to do some analysis tomorrow, but some outside analysis would also be helpful. | ||
| + | |||
| + | |||
| + | |||
| + | h4. Charles | ||
| + | |||
| + | |||
| + | |||
| + | FACS of various things. CHO Experiment from Tyler had very unhealthy cells. Kens cocultures have yet to be analyzed. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 20 | ||
| + | |||
| + | |||
| + | |||
| + | Divya - Miniprepped, Nanodropped, Inoculated. See Personal Notebook for details. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 19 | ||
| + | |||
| + | |||
| + | |||
| + | Divya - Transformed, Inoculated. See Personal Notebook for details. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 18 | ||
| + | |||
| + | |||
| + | |||
| + | Charles - FACS 100 samples in just under 90 minutes. Tyler's stuff had low concentrations so it ran slow, but data looks promising. Ken did Heks and Chos and both look promising. My stuff failed due to higher than intended initial cell concentration. Basically cells overgrew ran out of nutrients got sick and failed. Also made 20,000 ng of Dest 4-5.&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Tyler \-&nbsp;The NCAD failed...so far. It's only been 24 hours after transfection, but I didn't see any EYFP or "clumping". I know that my transfection efficiency isn't zero because I also transfected some CHO cells on the same plate with Hef1A:EBFP2 and wew saw about 5-10% efficiency. I will check again tomorrow (48 hrs) using the microscope and then send them for FACS. Maybe it was a bad batch of DNA... | ||
| + | |||
| + | |||
| + | |||
| + | Other things I did today: | ||
| + | |||
| + | |||
| + | |||
| + | \-FACS for TRE:LacI and TRE:LacI-Krab (I'm generating graphs now) | ||
| + | |||
| + | |||
| + | |||
| + | \-Transfected CHO cells for Notch-Delta Co-culture | ||
| + | |||
| + | |||
| + | |||
| + | \-I was going to repeat Mariola's Mnt and CI434 activation experiments but I couldn't find the DNA, so I asked Divya to do some LR reactions | ||
| + | |||
| + | |||
| + | |||
| + | The NCAD failed...so far. It's only been 24 hours after transfection, but I didn't see any EYFP or "clumping". I know that my transfection efficiency isn't zero because I also transfected some CHO cells on the same plate with Hef1A:EBFP2 and wew saw about 5-10% efficiency. I will check again tomorrow (48 hrs) using the microscope and then send them for FACS. Maybe it was a bad batch of DNA... | ||
| + | |||
| + | Other things I did today: | ||
| + | |||
| + | \-FACS for TRE:LacI and TRE:LacI-Krab (I'm generating graphs now) | ||
| + | |||
| + | \-Transfected CHO cells for Notch-Delta Co-culture | ||
| + | |||
| + | \-I was going to repeat Mariola's Mnt and CI434 activation experiments but I couldn't find the DNA, so I asked Divya to do some LR reactions | ||
| + | |||
| + | \-Tyler | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 17 | ||
| + | |||
| + | |||
| + | |||
| + | Jon - Prepped Notch for full sequencing, minipreps, cell stocking, nanodropping, prepping trash for pickup. | ||
| + | |||
| + | |||
| + | |||
| + | Divya - Inoculated and LR'd NCADs, Nanodropped CPLRs. Will transform tomorrow. See personal notebook for details. | ||
| + | |||
| + | |||
| + | |||
| + | h3. August 16 | ||
| + | |||
| + | |||
| + | |||
| + | Tiffany: Public wiki work forever. Don't use IE please. | ||
| + | |||
| + | |||
| + | |||
| + | Divya/Jon - In lab until 2am doing 26 minipreps, nanodrops, cell stocks, cleaning. :'( | ||
| + | |||
| + | |||
| + | |||
| + | Charles: Transfected cells with&nbsp;[EXP1-CH|iGEM2011:EXP1-CH]. Made graphs. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | h3. August 15 | ||
| + | |||
| + | |||
| + | |||
| + | Tyler: Prepared for transfections and updated the wiki. Will prepare CHO cells for NCAD phenotype experiment tomorrow. NCAD with HEK-293 cells failed (see&nbsp;[iGEM2011:NCAD Phenotype Verification]). Hef1A:GV16 experiments looked very good (see&nbsp;[Hef1A_GV16 Experiment|iGEM2011:Hef1A_GV16 Experiment]). TRE:LacI-EYFP-FF4 shows a lot of EYFP, about 25% increase if I remember correctly, but the LacI repression looks very weak. (see [TRE_LacI.Krab Experiments|iGEM2011:TRE_LacI.Krab Experiments])&nbsp;We have a Hef1A:mKate that is bad, showing about 0-2.5% fluorescence. I will repeat these experiments tomorrow. The DRD2 shows very small levels of activation, but I think it is actually there (see&nbsp;[iGEM2011:GPCR Experiments]). Plan for tomorrow's transfections: 1.)TRE:LacI-Krab 2.)TRE:LacI 3.)miRFF4 4.)Lipo Ladder. Wednesday: CHO transfections. | ||
| + | |||
| + | |||
| + | |||
| + | More thoughts: The GV16 and LacI-Krab experiments were done on the same day. I got about 35% efficiency from the GV16 experiment. The LacI-EYFP-FF4 is showing about 20% when activated. I think the leakiness of rtTA3 is skewing our picture, so in the next round of LacI experiments, I will also include \+/\- Hef1A:rtTA3 as an added control.&nbsp; | ||
| + | |||
| + | |||
| + | |||
| + | Kenneth: My apologies, but I can't make the morning meeting. Made the line chart for TRE:eBFP experiment:&nbsp;[TRE_eBFP2 verification|iGEM2011:TRE_eBFP2 verification]&nbsp;looks good. Processed data from TRE:mKate experiment:&nbsp;[TRE_mKate verification|iGEM2011:TRE_mKate verification]. Looks weird. No increase in red, but increase in blue, almost like TRE:eBFP2. Not quite sure what went wrong here. Maybe I aliquoted the wrong DNA, maybe there's some mixup? Either way, has to be redone. Future directions: continue working on the matlab code, process FACS data for co-cultures and AVPR2 testing as well as free Delta (all done Sunday). Experiments planned: | ||
| + | |||
| + | |||
| + | |||
| + | 1) Elowitz CHO cells [Initial CHO images - No Fluorescence]&nbsp;(so the cells don't express Delta-mCHerry without dox.) will FACS all of this Wednesday with proper scatter gating for CHO's | ||
| + | |||
| + | |||
| + | |||
| + | a. Dox induction of Delta (get a good curve showing conc of dox to amt of delta on surface | ||
| + | |||
| + | |||
| + | |||
| + | b. Addition of Free Delta to Notch-Gal4 cells and Notch+Delta cells | ||
| + | |||
| + | |||
| + | |||
| + | c. Co-culture with varying dox levels | ||
| + | |||
| + | |||
| + | |||
| + | d. If we see successful dox induction of Delta-mCherry and delta induction of Notch-Gal4/UAS, we can do the 2D ladder experiment where we vary both dox and delta levels for the Notch+Delta CHOs and see how much citrine, thus giving us a "surface" | ||
| + | |||
| + | |||
| + | |||
| + | 2)redo TRE:Delta-mCherry dox ladder-GET GOOD DATA THIS TIME | ||
| + | |||
| + | |||
| + | |||
| + | 3)TRE:eYFP and TRE:mKate need to be redone, I don't know if Charles wants to do this or not... | ||
| + | |||
| + | |||
| + | |||
| + | 4)Immunostaining using no-coverslip technique for pdisplay-vasopressin as well as AVPR2 for troubleshooting | ||
| + | |||
| + | |||
| + | |||
| + | 5)still waiting on free vasopressin for the AVPR2 tests. | ||
| + | |||
| + | |||
| + | |||
| + | 6)test HRH4 with histamine | ||
| + | |||
| + | |||
| + | |||
| + | 7)siRNA knockdown of N-cad. | ||
| + | |||
| + | |||
| + | |||
| + | Divya: Restriction Digested Clara's LRs. Send samples for sequencing. Labeled pRESC tubes. Got items from Weiss Lab. | ||
| + | |||
| + | |||
| + | |||
| + | Grant: Was amused at the bickering above. Designed quite a few primers\- this time around, for a Tango redesign and colored transactivators. Digested and gel extracted Sam's 4xFF4 gene entry backbone, pushing the Mnt-VP16-4xFF4 and Delta-mCherry-4xFF4 assemblies forward a bit. DNA work seems to have slowed considerably over the past few days\- we should make more (non-Tango) LRs. Ephrin stuff will hopefully be verified tomorrow. Takers? | ||
| + | |||
| + | |||
| + | |||
| + | Charles: Passed cells. Doing bunch of wiki work now. | ||
| + | |||
| + | |||
| + | |||
| + | Jon - Miniprepped and nanodropped 19 samples. Prepped SDMs for sequencing. Analyzed odd sequence results. Reinoculated Rescue 28 plasmids; put into incubator at 5:40pm, although many are already at high OD (wildly inconsistent though) so I'd check after a couple hours to make sure the E. coli don't reach stationary phase. | ||
<!----------------------------------------------------------------------------------------------------------------> | <!----------------------------------------------------------------------------------------------------------------> | ||
Revision as of 20:49, 28 September 2011
Overview
Here you can see what we did weekly in the lab!
Week 1: June 6 - June 11
In the first week we began constructing basic reporter DNA constructs that have various combinations of promoters and genes. The mix-and-matching of promoters and genes was done using the LR reaction following the Gateway protocol. Our promoters included TRE, minCMV, Hef1a, Hefl1a-LacO, and our genes included eYFP, mKate, and eBFP2. Transformed and inoculated colonies from the LR reactions and performed restriction digests. Approximately half of the LR reactions were successful and became DNA parts that we could use later on.June 7, 2011
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
C1434VP16 |
Yes. 6.7.2011, 5pm |
11 |
5, AMP, KH |
Standard, 30C for 16h |
| 6.7.2011 |
Michelle Louis |
2-3 |
Tre |
Mnt1VP16 |
Yes. 6.7.2011, 5pm |
12 |
4, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Tyler |
2-3 |
Tre |
LacIKrab |
Yes. 6.7.2011, 5pm |
6 |
30, AMP, KH | Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
3-4 |
minCMV-7xMnt1 |
eYFP |
Yes. 6.7.2011, 5pm |
9 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 |
Jenny Divya |
2-3 |
Tre |
LexAVP16 |
Yes. 6.7.2011, 5pm |
10 |
2, AMP, KH, needs to be redone |
Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 |
minCMV 1xC1434 |
eYFP |
Yes. 6.7.2011, 5pm |
7 |
3, AMP, KH, needs to be redone | Standard, 30C for 16h |
| 6.7.2011 | Mariola Kenneth |
3-4 | minCMV 4xLexA |
eYFP |
Yes. 6.7.2011, 5pm |
8 |
0, AMP, KH, needs to be redone |
Standard, 30C for 16h |
LR Reactions
Utilizing protocol here: LR Protocol
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Tube Number |
Colony Number, Antibiotic, Person |
Protocol Used |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Grant |
3-4 |
minCMV 4xLexA | eYFP | Yes. 6.9.2011, 3pm |
12 |
10, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xMnt1 |
eYFP |
Yes. 6.9.2011, 3:45pm | 2 |
13, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
minCMV-4xCI434 |
eYFP |
Yes. 6.9.2011, 3:45pm | 1 |
7, AMP, GR | Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Jon C |
3-4 |
Hef1a-LacO |
eYFP |
Yes, 6.9.2011, 4:20pm |
5 |
25, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
4-5 |
Tre |
mKate |
Yes, 6.9.2011 4:27pm |
3 |
6, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
| 6.9.2011 |
Semon |
1-2 |
Hef1a |
eBFP2 |
Yes, 6.9.2011, 4.27pm |
4 |
3, AMP, GR |
Standard + Grown at 37C in LB not SOC. |
Transformation
| Date |
Assignee |
DEST_R4R2 |
L4-R1 Promoter |
L1-L2 Gene |
LR? |
Colony Number, Antibiotic |
Protocol Used | |
|---|---|---|---|---|---|---|---|---|
| 6.9.2011 |
Mariola |
|
|
eYFP |
no. Replication stock. | |
2, AMP |
Transformation (~ 2 hr 20 min) using LB instead of SOC |
Miniprep
| Date | Assignee |
DNA |
Quantity |
Time collected |
|---|---|---|---|---|
| 6.9.2011 |
Divya-Jenny |
pEXPR_2-3_Tre:LexAVP16 |
52.2 ng/uL |
12pm |
| 6.9.2011 |
Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A) |
70 ng/ul |
12pm |
| 6.9.2011 |
Kenneth | pEXPR_3-4_minCMV-CI434:eYFP (B) |
174 ng/uL |
12pm |
| 6.9.2011 |
Louis |
pEXPR_2-3_Tre:C1434VP16 |
234.6 ng/uL |
12pm |
| 6.9.2011 |
Tyler |
pEXPR_2-3_Tre:Lac/Krab |
130.7 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
110.0 ng/uL |
12pm |
| 6.9.2011 |
Michelle |
pDEST_2-3_ccdB |
117.6 ng/uL |
12pm |
Restriction Digests
| Assignee |
DNA |
Enzyme |
Expected Results |
Picture of Gel |
Time Incubated | Comments |
||
|---|---|---|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | NdeI |
6700 bp 800 bp |
a.3 | 6/9/11 3:50 PM |
|
||
| Kenneth |
pEXPR_3-4_minCMV-CI434:eYFP (A+B) |
NcoI and SacII |
2450 bp 4650 bp |
A: a.6 B: a.7 |
6/9/11 3:00 PM |
A did not cut as expected. B looks good. Digestion Mix: 2 uL NEB4, 0.5 uL of each enzyme, A: 7.1 uL/B: 2.9 uL of DNA, 10.4 uL/14.6 uL of H20 |
||
| Michelle |
pEXPR_2-3_Tre:Mnt1VP16 |
BglI |
3700 bp 2200 bp 1300 bp |
a.1 | 6/9/11 4:00PM |
DNA appeared to be uncut. Need to redo, possibly with a new enzyme and more DNA. |
||
| Michelle |
pDEST_2-3_ccdB |
NcoI and NheI |
6100 bp 1700 bp |
a.2 | 6/9/2011 4:25 PM |
Attained bands at correct positions. Other band represents partially cut DNA in double digest. |
||
| Divya |
pEXPR_2-3_Tre:LexAVP16 |
HincII |
a.4 | 6/9/2011 4:45 PM |
- Bands didn't show up. Needs to be redone. - DNA may have degraded. Need to redo Nanodrop as well. |
|||
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
a.5 | 6/9/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL Only saw one band at 3500 bp Needs to be redone possibly with another restriction enzyme |
Gels
June 10, 2011
Restriction Gels
| Assignee | DNA |
Enzyme |
Expected result |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Louis |
pEXPR_2-3_Tre:C1434VP16 | EcoRI (Buffer 2) |
5500 bp 1050 bp 650 bp 360 bp |
6/10/11 11:15 AM |
|
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (A) |
NcoI and NheI |
5700 bp 1300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NcoI 1 uL Nhe1 2 uL NEB2 2 uL BSA 8 uL H2O Failed. |
| Michelle |
pEXPR_2-3_TRE:Mnt1VP16 (B) |
NheI and SphI |
1700 bp 5300 bp |
6/10/11 11:40 AM |
Digestion: 6 uL DNA 1 uL NheI 1 uL SphI 2 uL NEB2 2 uL BSA 8 uL H2O Failed. Redoing LR on Monday June 13th. |
| Tyler | pEXPR_2-3_Tre:Lac/Krab | SalI | 3 bands: 5288 bp 1510 bp 1241 bp cuts gene |
6/10/2011 3:00 PM |
Digestion: 4 uL * 130.7 ng/uL = 522.8 ng DNA 2 uL NEB3 2 uL BSA 11 uL H2O 1 uL SalI Total: 20 uL ***Worked (see gel below - b.1) Stored in -80C freezer |
Comments:
pEXPR_2-3_Tre:LexAVP16 and pEXPR_3-4_minCMV-7xMnt1:eYFP are being entirely redone.
pEXPR_2-3_Tre:LexAVP16 (post-miniprep) was lost. pEXPR_3-4_minCMV-7xMnt1:eYFP LR was unsuccessful (no bacteria grew).
LR and transformation will be done on Saturday. Miniprep and restriction mapping will be done on Sunday
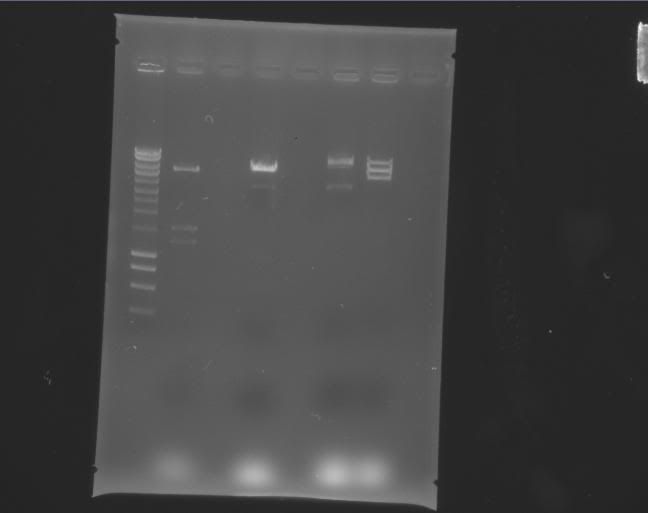
| Label | Gel |
Legend |
|---|---|---|
| b |
 > > |
Column 0: HyperLadder Column 1: pEXPR_2-3_Tre:Lac/Krab Column 4: pEXPR_2-3_Tre:C1434VP16 Column 6: pEXPR_2-3_TRE:Mnt1VP16 (A) Column 7: pEXPR_2-3_TRE:Mnt1VP16 (B) |
June 11, 2011
Set of LRs from 6/9 (GR, JC, and SR) cell counts taken; inoculated by GR/JC. 3 cells taken per plate.
| Assignee |
DNA |
Enzyme |
Expected Results |
Time Incubated |
Comments |
|---|---|---|---|---|---|
| Sam |
pEXPR_1-2_Hef:eBFP |
NcoI & ApaL1 |
3750bp, 3180bp, 1250bp | 45 min | If it doesn't work there should be a whole mess of fragments (including a small 600bp one) 1mL each enz; 2 mL Buf; 1 mL BSA; 2mL DNA; 13mL H2O (Standard Protocol with extra BSA) |
| Sam |
pEXPR_4-5_Tre:mKate |
Bgl1 |
1270bp, 2630bp, 3380bp | 45 min | 1mL enz; 2mL DNA; 2mL BSA; 2mL Buf; 15mL H2O (Standard Protocol with extra BSA and water - not quite 10x) |
 "
"