Team:NYC Software/Deinococcus/Data
From 2011.igem.org
| Line 15: | Line 15: | ||
<div id="container"> | <div id="container"> | ||
| + | <h4>What we have learned about the Deinococcus Genera</h4> | ||
| + | <h4>Different Phylogenetic Methods Yield Different Clades</h4> | ||
| + | |||
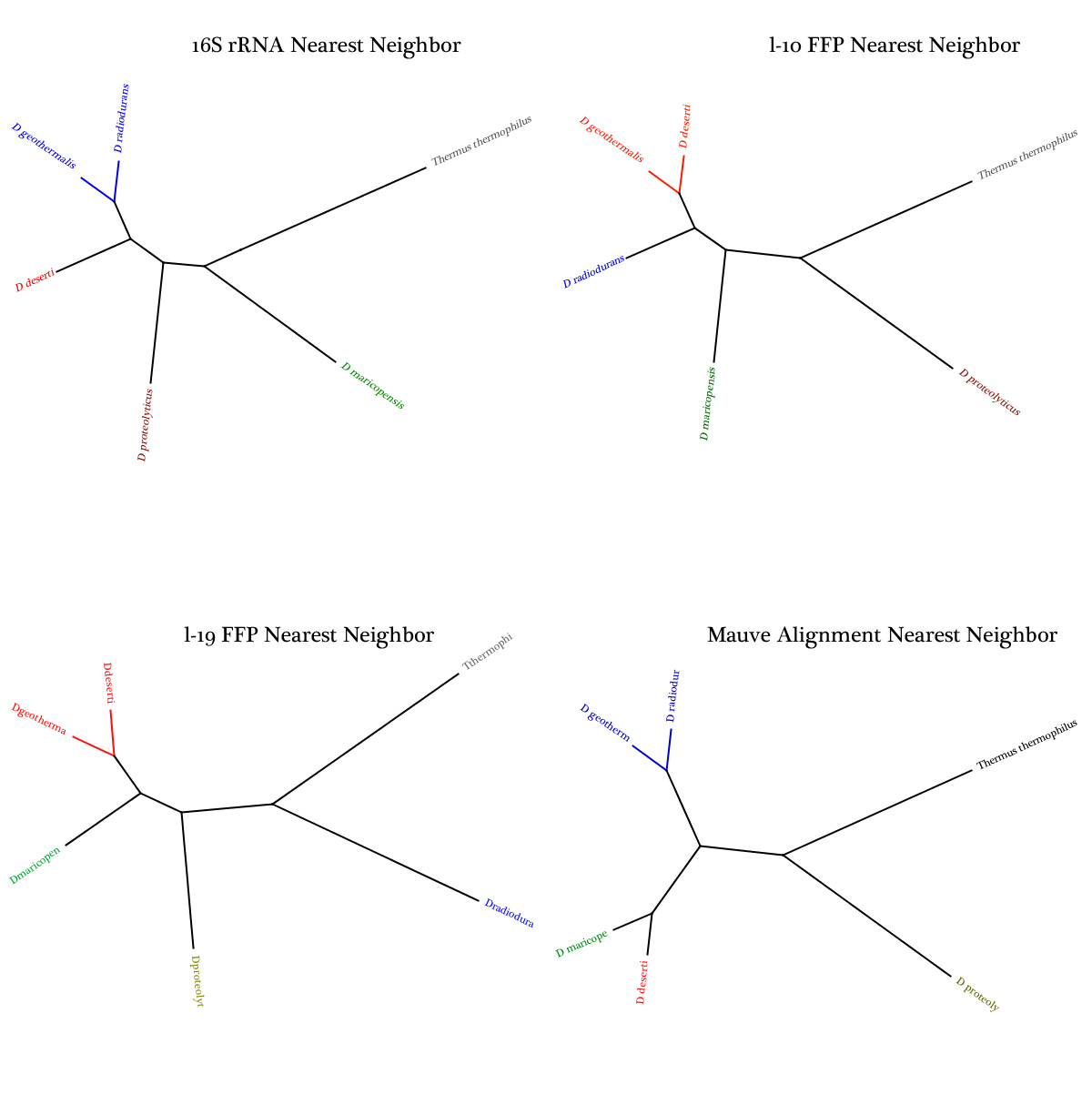
| + | We extrapolated the evolutionary relationships between the species using 1) Feature Frequency Profile comparisons, 2) 16S rRNA alignments, and 3) Mauve genome alignment, but each method yielded an alternative result. | ||
| + | <br>By sequencing more species in this family, we hypothesize that Inclusion of a larger set of sequenced species in the genera may elucidate new and more reliable information about their origins and cladistics. | ||
| + | <p> | ||
| + | <strong>Disparate Trees Resulting From Too Little Genetic Information</strong> | ||
| + | |||
| + | |||
| + | <img src='https://static.igem.org/mediawiki/2011/e/e8/Phylogeny_comparison.jpg' align='left' width=650px> | ||
| + | |||
<h2> Genome Analysis </h2> | <h2> Genome Analysis </h2> | ||
Latest revision as of 00:36, 29 September 2011

What we have learned about the Deinococcus Genera
Different Phylogenetic Methods Yield Different Clades
We extrapolated the evolutionary relationships between the species using 1) Feature Frequency Profile comparisons, 2) 16S rRNA alignments, and 3) Mauve genome alignment, but each method yielded an alternative result.By sequencing more species in this family, we hypothesize that Inclusion of a larger set of sequenced species in the genera may elucidate new and more reliable information about their origins and cladistics.
Disparate Trees Resulting From Too Little Genetic Information
Genome Analysis
While waiting for the sequencing data from our libraries, we started working with the 5 already sequenced genomes in the family. Because we are most interested in finding potential radiation-resistant genes to help support the NYC_Wetware Team's data, we wanted to build a pipeline capable of predicting genes in the species, comparing those genes to find the most similar ones and then characterizing their relationships by probing radiation-resistance pathways for unique conservation.
To identify the most related genes, we predicted genes with Glimmer3 and then aligned those reads to the Deinococcus radiodurans genome. For each CDS in the D. rad genome, a number of related genes from the various species is aligned. At the time of the wiki freeze we have alignment files detailing these sequence relationships and are working toward adding biological pathway information.

 "
"