Team:Lethbridge/Attributions
From 2011.igem.org
Liszabruder (Talk | contribs) (→Attributions & Contributions) |
Liszabruder (Talk | contribs) |
||
| Line 35: | Line 35: | ||
The University of Lethbridge iGEM team would like to acknowledge the following people for their attributions and expertise which added to the success of our project, it is greatly appreciated, and has helped us gain new knowledge and skills. | The University of Lethbridge iGEM team would like to acknowledge the following people for their attributions and expertise which added to the success of our project, it is greatly appreciated, and has helped us gain new knowledge and skills. | ||
| - | ==Dr. A. William Smith== | + | ===Dr. A. William Smith=== |
from The Department of New Media at the University of Lethbridge – For his assistance, knowledge, leadership and hard work, in the Biospirits film project. | from The Department of New Media at the University of Lethbridge – For his assistance, knowledge, leadership and hard work, in the Biospirits film project. | ||
| - | ==Mr. Doug Bray== | + | ===Mr. Doug Bray=== |
from The Faculty of Arts and Science at the University of Lethbridge – For his training and guidance of team members in the use of transmission electron microscopy. | from The Faculty of Arts and Science at the University of Lethbridge – For his training and guidance of team members in the use of transmission electron microscopy. | ||
| - | ==Dr. Mark Roussel== | + | ===Dr. Mark Roussel=== |
from The Department of Chemistry and Biochemistry at the University of Lethbridge – For his assistance with protein modeling. | from The Department of Chemistry and Biochemistry at the University of Lethbridge – For his assistance with protein modeling. | ||
| - | ==Mr. David Lain Huston and the Arnott labs== | + | ===Mr. David Lain Huston and the Arnott labs=== |
from The Institute of Cellular, Molecular, and Systems biology in Glasgow – For sending the team genes from the Xylene degradation pathway. | from The Institute of Cellular, Molecular, and Systems biology in Glasgow – For sending the team genes from the Xylene degradation pathway. | ||
| Line 51: | Line 51: | ||
=Collaboration - Calgary iGEM= | =Collaboration - Calgary iGEM= | ||
We were asked by the Calgary iGEM team to to test the viability of their chassis to survive in tailings pond water. If <i>E. coli</i> DH5α cells containing one of our constructs can grow normally in medium prepared with tailings water, we can be confident that the chassis can be applied to tailings pond water in general. | We were asked by the Calgary iGEM team to to test the viability of their chassis to survive in tailings pond water. If <i>E. coli</i> DH5α cells containing one of our constructs can grow normally in medium prepared with tailings water, we can be confident that the chassis can be applied to tailings pond water in general. | ||
| - | ==Materials and Methods== | + | ===Materials and Methods=== |
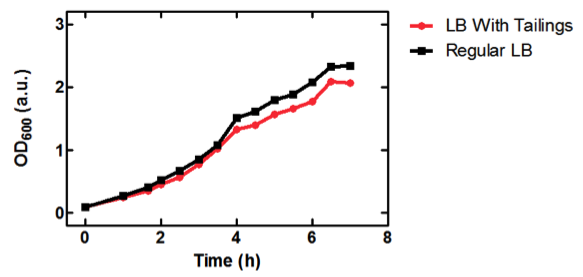
50 mL of LB medium was made with tailings water and filtered through MILLEX filter Unit GS MF-Millipore MCE Membrane 0.22 µm in order to sterilize it and appropriate antibiotics were added. The tailings LB was inoculated with <i>E. coli</i> DH5α cells containing BBa_K331009 to an OD<sub>600</sub> of 0.1. The flask was incubated at 37˚C with shaking for 7 hours, during which photometric readings were taken every 30. Photometric readings were taken using the Pharmacia Biotech Ultrospec 3000, as the OD<sub>600</sub> readings reached 1.0 the solutions was diluted with LB media to keep the OD<sub>600</sub> reading between 0.1 and 1.0. The same protocol was used to observe cell growth of <i>E. coli</i> DH5α cells containing BBa_K331009 in 50 mL of LB medium made with MilliQ H<sub>2</sub>O. | 50 mL of LB medium was made with tailings water and filtered through MILLEX filter Unit GS MF-Millipore MCE Membrane 0.22 µm in order to sterilize it and appropriate antibiotics were added. The tailings LB was inoculated with <i>E. coli</i> DH5α cells containing BBa_K331009 to an OD<sub>600</sub> of 0.1. The flask was incubated at 37˚C with shaking for 7 hours, during which photometric readings were taken every 30. Photometric readings were taken using the Pharmacia Biotech Ultrospec 3000, as the OD<sub>600</sub> readings reached 1.0 the solutions was diluted with LB media to keep the OD<sub>600</sub> reading between 0.1 and 1.0. The same protocol was used to observe cell growth of <i>E. coli</i> DH5α cells containing BBa_K331009 in 50 mL of LB medium made with MilliQ H<sub>2</sub>O. | ||
| - | ==Results== | + | ===Results=== |
[[image:uofltailings.png|center|400px]] | [[image:uofltailings.png|center|400px]] | ||
<b>Figure 1.</b> OD<sub>600</sub> of <i>E. coli</i> DH5α cells containing BBa_K331009 cultured in LB medium made with tailings water (red) and LB medium made with MilliQ H<sub>2</sub>O (black). | <b>Figure 1.</b> OD<sub>600</sub> of <i>E. coli</i> DH5α cells containing BBa_K331009 cultured in LB medium made with tailings water (red) and LB medium made with MilliQ H<sub>2</sub>O (black). | ||
| - | ==Conclusion== | + | ===Conclusion=== |
As seen in Figure 1, <i>E. coli</i> DH5α cells were able to grow in LB medium made with tailings water, suggesting that the chassis will survive for use in tailings pond water. | As seen in Figure 1, <i>E. coli</i> DH5α cells were able to grow in LB medium made with tailings water, suggesting that the chassis will survive for use in tailings pond water. | ||
<br> | <br> | ||
<br> | <br> | ||
Revision as of 19:37, 28 September 2011
|
|
|
|---|
 "
"