Team:Harvard/Technology
From 2011.igem.org
| Line 22: | Line 22: | ||
[https://2011.igem.org/Team:Harvard/Results/MAGE MAGE results] | [https://2011.igem.org/Team:Harvard/Results/MAGE MAGE results] | ||
| - | |||
| - | |||
[https://2011.igem.org/Team:Harvard/ZF_Binding_Site_Finder ZFP binding site finder] | [https://2011.igem.org/Team:Harvard/ZF_Binding_Site_Finder ZFP binding site finder] | ||
Revision as of 00:31, 23 September 2011
All subpage links go here (Temporary):
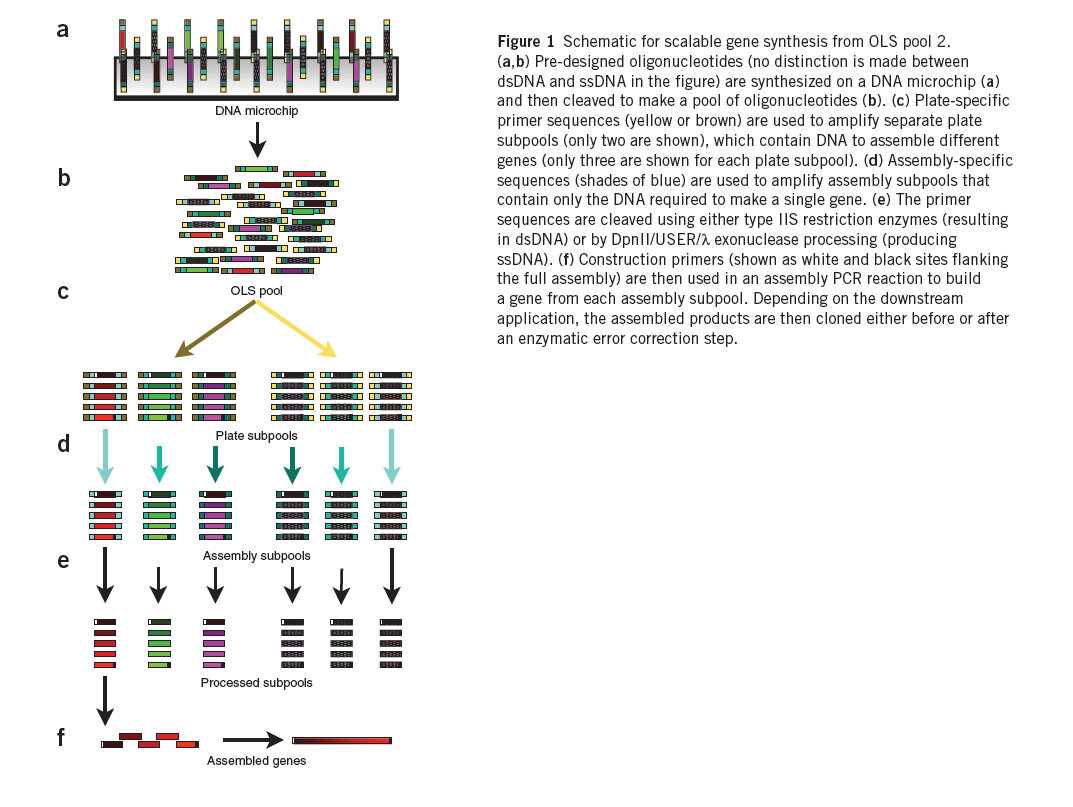
Chip-Based DNA Synthesis
We are creating 55,000 zinc fingers using microchip synthesis (Kosuri et al). These fingers will then be tried against the DNA sequences we wish to bind.
Summary (Adapted from Kosuri et al)1
The synthesis of DNA encoding regulatory elements, genes, pathways and entire genomes provides powerful ways to both test biological hypotheses and harness biology for our use. For example, from the use of oligonucleotides in deciphering the genetic code to the recent complete synthesis of a viable bacterial genome, DNA synthesis has engendered tremendous progress in biology. Currently, almost all DNA synthesis relies on the use of phosphoramidite chemistry on controlled-pore glass (CPG) substrates. The synthesis of gene-sized fragments (500–5,000 base pairs (bp)) relies on assembling many CPG oligonucleotides together using a variety of gene synthesis techniques. Technologies to assemble verified gene-sized fragments into much larger synthetic constructs are now fairly mature.
The price of gene synthesis has fallen drastically over the last decade. However, the current commercial price of gene synthesis, ~$0.40–1.00/bp, has begun to approach the relatively stable cost of the CPG oligonucleotide precursors (~$0.10–0.20/bp)1, suggesting that oligonucleotide cost is limiting. At these prices, the construction of large gene libraries and synthetic genomes is out of reach to most.
A promising route is to harness existing DNA microchips, which can produce up to a million different oligonucleotides on a single chip, as a source of DNA. Recently, the quality of microchip-synthesized oligonucleotides was improved by controlling depurination during the synthesis process. These arrays produce up to 55,000 200-mer oligonucleotides on a single chip and are sold as a ~1–10 picomole pools of oligonucleotides, termed OLS pools (oligo library synthesis). Estimations of the frequency of transitions, transversions, insertions and deletions in OLS pools found the overall error rate to be ~1/500 bp both before and after PCR amplification, suggesting that OLS pools can be used for accurate large-scale gene synthesis.
MAGE
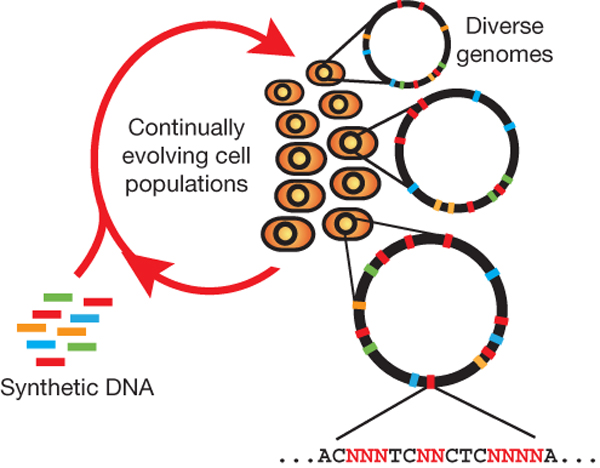
Summary (Adapted from Wang et al)2,3
The breadth of genomic diversity found among organisms in nature allows populations to adapt to diverse environments. However, genomic diversity is difficult to generate in the laboratory and new phenotypes do not easily arise on practical timescales. Although in vitro and directed evolution methods have created genetic variants with usefully altered phenotypes, these methods are limited to laborious and serial manipulation of single genes and are not used for parallel and continuous directed evolution of gene networks or genomes.
Multiplex automated genome engineering (MAGE) is a new method for large-scale programming and evolution of cells. MAGE simultaneously targets many locations on the chromosome, thus producing combinatorial genomic diversity. Because the process is cyclical and scalable, MAGE facilitates rapid and continuous generation of a diverse set of genetic changes (mismatches, insertions, deletions). This multiplex approach embraces engineering in the context of evolution by expediting the design and evolution of organisms with new and improved properties.
MAGE provides a highly efficient, inexpensive and automated solution to simultaneously modify many genomic locations (for example, genes, regulatory regions) across different length scales, from the nucleotide to the genome level.
Lambda Red- Mediated Recombineering
Useful Links
- Gene Knockouts and Exchanges by Linear Transformation: http://rothlab.ucdavis.edu/protocols/Lin.Transform.html
- Open Wet Ware Protocol: http://openwetware.org/wiki/Recombineering/Lambda_red-mediated_gene_replacement
Gibson (Isothermal) Assembly
Summary (Adapted from Gibson et al)4,5
- Source: http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html
- Protocol: http://www.nature.com/protocolexchange/protocols/554#/main
References
1. Sriram Kosuri, Nikolai Eroshenko, Emily M LeProust, Michael Super, Jeffrey Way, Jin Billy Li, George M Church. (2010). Scalable gene synthesis by selective amplification of DNA pools from high-fidelity microchips. Nature Biotechnology, 28(12):1295-9. [http://www.nature.com/nbt/journal/v28/n12/full/nbt.1716.html]
2. Harris H. Wang, Farren J. Isaacs, Peter A. Carr, Zachary Z. Sun, George Xu, Craig R. Forest, George M. Church. Programming cells by multiplex genome engineering and accelerated evolution. (2009). Nature, 460(7257):894-8. [http://www.nature.com/nature/journal/v460/n7257/full/nature08187.html]
3. Isaacs FJ, Carr PA, Wang HH, Lajoie MJ, Sterling B, Kraal L, Tolonen AC, Gianoulis TA, Goodman DB, Reppas NB, Emig CJ, Bang D, Hwang SJ, Jewett MC, Jacobson JM, Church GM. (2011). Precise manipulation of chromosomes in vivo enables genome-wide codon replacement. Science, 333(6040):348-53. [http://www.sciencemag.org/content/333/6040/348.short]
4. Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, Smith HO. (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods,6(5):343-5. Epub 2009 Apr 12.
[http://www.nature.com/nmeth/journal/v6/n5/full/nmeth.1318.html]
5. Gibson D. (2009). One-step enzymatic assembly of DNA molecules up to several hundred kilobases in size. Nature Protocols,Published online 16 April 2009, doi:10.1038/nprot.2009.77.
[http://dx.doi.org/10.1038/nprot.2009.77]
 "
"