Commons
Miniprep
Investigators: Sandra
Yesterday LB was inoculated with strains (S14, S39-S44 etc.). Today miniprep of strains were performed.
Digestion: 2A-assembly
Investigators: Sandra
Digestion of:
with XbaI and PstI
- S39 (PR1) pSB1C3
- S40 (PR2) pSB1C3
- S41 (PR3) pSB1C3
- S42 (PR4) pSB1C3
- S43 (PR5) pSB1C3
- S44 (PR6) pSB1C3
with SpeI and PstI
incubation for 6 hours at 37°C.
Inactivation of enzymes at 80°C for 20 minutes.
Samples were loaded onto a gel and there were no inserts of GFP.
Therefore RFP (S1b, S2a in pSB1A3) was digested with SpeI and PstI over night to ligate it with PR1-PR6 tomorrow.
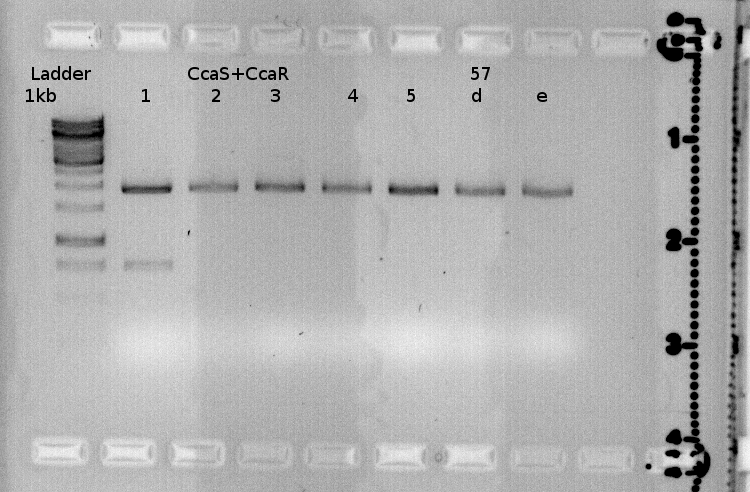
green light receptor
sequence analysis
Investigators:Julia
we had following sequencing results
46b= ho1 with a RBS (BBa_B0034), correct
50b= RBS(BBa_B0034)+pcyA+terminator, correct
64b= Promotor (BBa_J23110)+CcaS, wrong big part is missing
67b= Promotor (BBa_J23110)+CcaR, correct
we realised that there must have been a mistake during quickchange PCR of CcaS from 22.08.
The primer annealed in a unspecific way and because of that a big part of about 380 bp is missing.
Testdigest of 3A-assembly from 3.09

PCB is supposed to be the assembled gene for ho1 and pcyA.</br>
The expected bands for the insert are missing.</br>
ho1 is the gel-extracted PR-ho1+ terminator construct.</br>
The one sample with three bands will be send for sequencing.</br>
|
blue light receptor
Trafo
Investigators: Sandra
Trafo of ligated parts of the 2A-assembly.
Sequence analysis
sequence analysis of our lovtap-Notgate was neagtive:-( So we´ll try the gibson assembly.
PCR
Investigators: Sophie
PCR
| Name: Sophie
| Date: 06.09.11
|
| Continue from Experiment (Date)
(Name)
|
| Project Name: Gibson of ♥ and NOT
|
PCR-Mixture for one Reaction:
For a 50 µl reaction use
| 32,5µl
| H20
| Name
|
| 10µl
| 5x Phusion Buffer
| of Primer
|
| 2.5µl
| Primer fw
| ♥: P97
NOT: P99
|
| 2.5µl
| Primer dw
| ♥: P98
NOT: P100
|
| 1µl
| dNTPs
| of Template DNA
|
| 1µl
| DNA-Template
| NoT, M45 (BBa_Q0400)
original DNA from Edinburgh (BBa_K322999)
|
| 0.5 µl
| Phusion (add in the end)
|
|
Digestion with Dpn I and purification with PCR purification kit
What program do you use?
First 20 cycles touchdown 69°C -0.4/cycle
next 10 cycles touchdown 72°C -0.2 per cycle
PCR products were loaded onto a gel, but there were no bands execpt for not3.
Lysis cassette
Troubleshooting of the modified Lysis genes K124017
Investigators:Theo
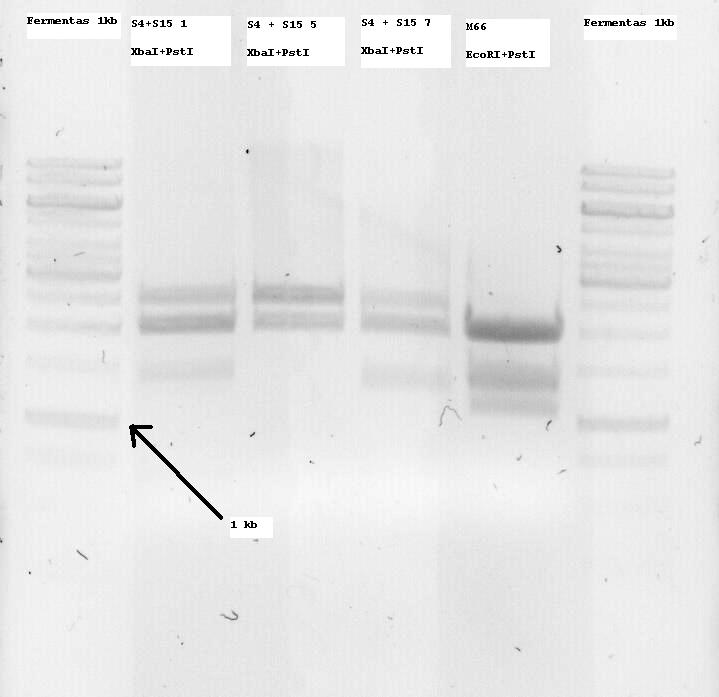
S4+S15 Nr1, 5 and 7 were sent for sequencing. Some problems arose and the sequences seemed to not be correct or only parts of them. So I decided to digest all of the minis and if an insert band (ie about 1300bp, since it is K124017 with RBS) was seen, to extract it from the gel and use it for subsequent ligation with the Promotor.
M66 which is (or SHOULD be -arrghh-) the already modified Lysis genes K124017 was digested with EcoRI and PstI and two bands at around 1100 and 1400bp were seen, so both of them were extracted. I want to clone them on the pSB1C3 Vector and sequence that so that the part is also ready for the parts registry. More on that tommorow.
- Note: S4 is the B0034 RBS (ribosome binding site), S15 is the non-modified K124017 Lysis Cassette without RBS
- Note 2: S4+S15 Nr1, 5 and 7 were digested with XbaI + PstI so that they can be one-step cloned with the temperature sensitive promotor (to be cut with SpeI and PstI and then treated with alcalic phosphatase as well as with the Qiagen PCR purification kit)
Gel Extraction Kit
Qiagen Kit
| Name: Theo
| Date: 6.9.2011
|
| Continue from Experiment: Troubleshooting of the modified Lysis genes K124017 (5.9.2011, Theo)
|
| Project Name: Troubleshooting of the modified Lysis genes K124017
|
Procedure
1.Excise the DNA fragment from the agarose gel with a clean, sharp scalpel.
2.Weigh the gel slice in a colorless tube. Add 3 volumes Buffer QG to 1 volume gel (100 mg ~ 100 μl). For >2% agarose gels, add 6 volumes Buffer QG.
3.Incubate at 50°C for 10 min (or until the gel slice has completely dissolved). Vortex the tube every 2–3 min to help dissolve gel.
4.After the gel slice has dissolved completely, check that the color of the mixture is yellow (similar to Buffer QG without dissolved agarose). If the color of the mixture is orange or violet, add 10 μl 3 M sodium acetate, pH 5.0, and mix. The color of the mixture will turn yellow.
5.Add 1 gel volume of isopropanol to the sample and mix.
6.Place a QIAquick spin column in ␣a provided 2 ml collection tube or into ␣a vacuum manifold.
7.To bind DNA, apply the sample to the QIAquick column and ␣centrifuge for 1 min or ␣apply vacuum to the manifold until all the samples have passed through the column.␣Discard flow-through and place the QIAquick column back into the same tube. For sample volumes of >800 μl, load and spin/apply vacuum again.
8.If the DNA will subsequently be used for sequencing, in vitro transcription, or microinjection, add 0.5 ml Buffer QG to the QIAquick column and␣centrifuge for 1 min or␣apply vacuum.␣Discard flow-through and place the QIAquick column back into the same tube.
9.To wash, add 0.75 ml Buffer PE to QIAquick column and ␣centrifuge for 1 min or␣apply vacuum.␣Discard flow-through and place the QIAquick column back into the same tube.
Note: If the DNA will be used for salt-sensitive applications (e.g., sequencing, blunt-ended ligation), let the column stand 2–5 min after addition of Buffer PE.
10.Centrifuge the QIAquick column once more in the provided 2 ml collection tube for 1 min at 17,900 x g (13,000 rpm) to remove residual wash buffer.
11.Place QIAquick column into a clean 1.5 ml microcentrifuge tube.
12.To elute DNA, add 50 μl Buffer EB (10 mM Tris·Cl, pH 8.5) or water to the center of the QIAquick membrane and centrifuge the column for 1 min. For increased DNA concentration, add 30 μl Buffer EB to the center of the QIAquick membrane, let the column stand for 1 min, and then centrifuge for 1 min. After the addition of Buffer EB to the QIAquick membrane, increasing the incubation time to up to 4 min can increase the yield of purified DNA.
13.If the purified DNA is to be analyzed on a gel, add 1 volume of Loading Dye to 5 volumes of purified DNA. Mix the solution by pipetting up and down before loading the gel.
Precipitator
Transformation
| Name: Rüdiger
| Date: 06.09. - 07.09
|
| Continue from Date 05.09. Name Rüdiger
Experiment Ligation
|
| Project Name: Precipitator
|
Procedure
- take cells from -80°C freezer and put them on ice! (every eppi contains about 400 μl cells)
- thaw cells on ice 20 minutes
- pipette 50 μl cells and 2 μl DNA into eppi still on ice!
- Incubate for 30 minutes on ice
- Heat at 42°C for 60 sec
- Incubate on ice for 5 minutes
- Add 200 μl LB Broth
- Incubate for 2 hours at 37°C (cells with lysis cassette at 30°C!!)
- Plate 50 μl and 200μl on two different LB/Agar plates with appropriate antibiotic resistance
Documentation:
Why are you doing this experiment? Name of the sample? Where are they stored? Name the vector with inserts, antibiotika resistance etc.
| Picked colonies from plates 1 - 10
|
 "
"
 Contact
Contact