Team:Potsdam Bioware/Labjournal/August part 1
From 2011.igem.org
| Line 5,462: | Line 5,462: | ||
** 1 plate with cm (25µg/ml), 1 mM IPTG, 20 mM ara<br> | ** 1 plate with cm (25µg/ml), 1 mM IPTG, 20 mM ara<br> | ||
| + | |||
| + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Survival test for E.coli XL1 blue transformed with pUP_SG2_ssTorA_CS-Pre_bla </h3> | ||
| + | |||
| + | <b>Investigator:</b> Sebastian, Stefan, Sascha<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <b>Aim:</b><br> | ||
| + | |||
| + | *testing the influence of the induction with IPTG and arabinose (different concentrations)<br> | ||
| + | |||
| + | *capacity of resistence vs. ampicillin after induction of TorA_CS-TEV_bla with IPTG<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <b>Materials:</b><br> | ||
| + | |||
| + | * LB-Agar<br> | ||
| + | |||
| + | * deluted overnight culture of E.coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV<br> | ||
| + | |||
| + | * LB-Media<br> | ||
| + | |||
| + | * Stock solutions of 1M IPTG, 1M arabinose (ara), 100mg/ml ampicillin (amp) and 25 mg/ml chloramphenicol (cm)<br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <b>Methode:</b> | ||
| + | |||
| + | 100µl of deluted overnight culture of E.coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV (OD (600 nm)=0.002) were plated on different perpared agar plates and incubated over night at 30°C.<br> | ||
| + | |||
| + | *<i>used plates:</i><br> | ||
| + | |||
| + | **total plates: 15<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml)<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG <br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG, amp (50 µg/ml)<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG, amp (100 µg/ml)<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG, amp (200 µg/ml)<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG, amp (400 µg/ml)<br> | ||
| + | |||
| + | ** 1 plate with cm (25µg/ml), 1 mM IPTG, amp (800 µg/ml)<br> | ||
| + | <b>Results:</b><br> | ||
| + | |||
| + | <br> | ||
| + | |||
| + | <b>Further tasks:</b><br> | ||
<b>Results:</b><br> | <b>Results:</b><br> | ||
Latest revision as of 01:03, 22 September 2011
50th Labday 2011-08-01
Sequencing of mutated mdnA genes
Investigators: Steffi, Vanessa, Nicole
Aim: Determination of mutation rate employing sequencing of mutated mdnA
Time: 2011-08-01,10:00-13:00
Materials:
- Miniprep of mutated mdnA and restricted for 2 resp. 3 hours
- Sequencing Primer: sf_mdna_1
- Freelabels for Value ReadTube (MWG Eurofins)
Method:
- DNA concentration (for sequencing): 100 ng/ µl
- Total volume: 15 µl
- Primer concentration: 2 pmol/ µl
- Total volume: 200 µl (approx. 15 µl per sequencing reaction)
- sent to MWG Eurofins with the aid of Free sample bags
Further tasks:
- Analyzing sequencing results
- Determination of mutation rates
Ligation of mdnA and GeneIII for phage display (strategy 2)
Investigators: Leif
Aim:ligation of mdnA and GenIII to get a fusion gene for phage display
Time: 11:00-13:00
Method:
- ligation-samples from 2011-07-27;
Protocol:
- 5 µl (ca 25 ng) digested geneIII (NgoMIV, AatII)
- 3 µl (ca 30 ng) digested mdnA (NarI, AgeI)
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 9 µl water
2 h, room temperature
Further tasks:
ligation into vector
Ligation of mdnA/GeneIII-fusion gene into pARW089 for phage display (strategy 2)
Investigators: Leif
Aim: ligation of mdnA and GenIII to get a fusion gene for phage display
Time: 11:00-13:00
Method:
- ligation-samples from 2011-08-01;
Protocol:
- 6 µl (ca 70 ng) digested vector pARW089 (NarI, AatII)
- 1 µl (ca 5 ng) fusion gene mdnA/geneIII
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 10 µl water
2 h, room temperature, then over night in the freezer
Further tasks:
transformation of E. coli
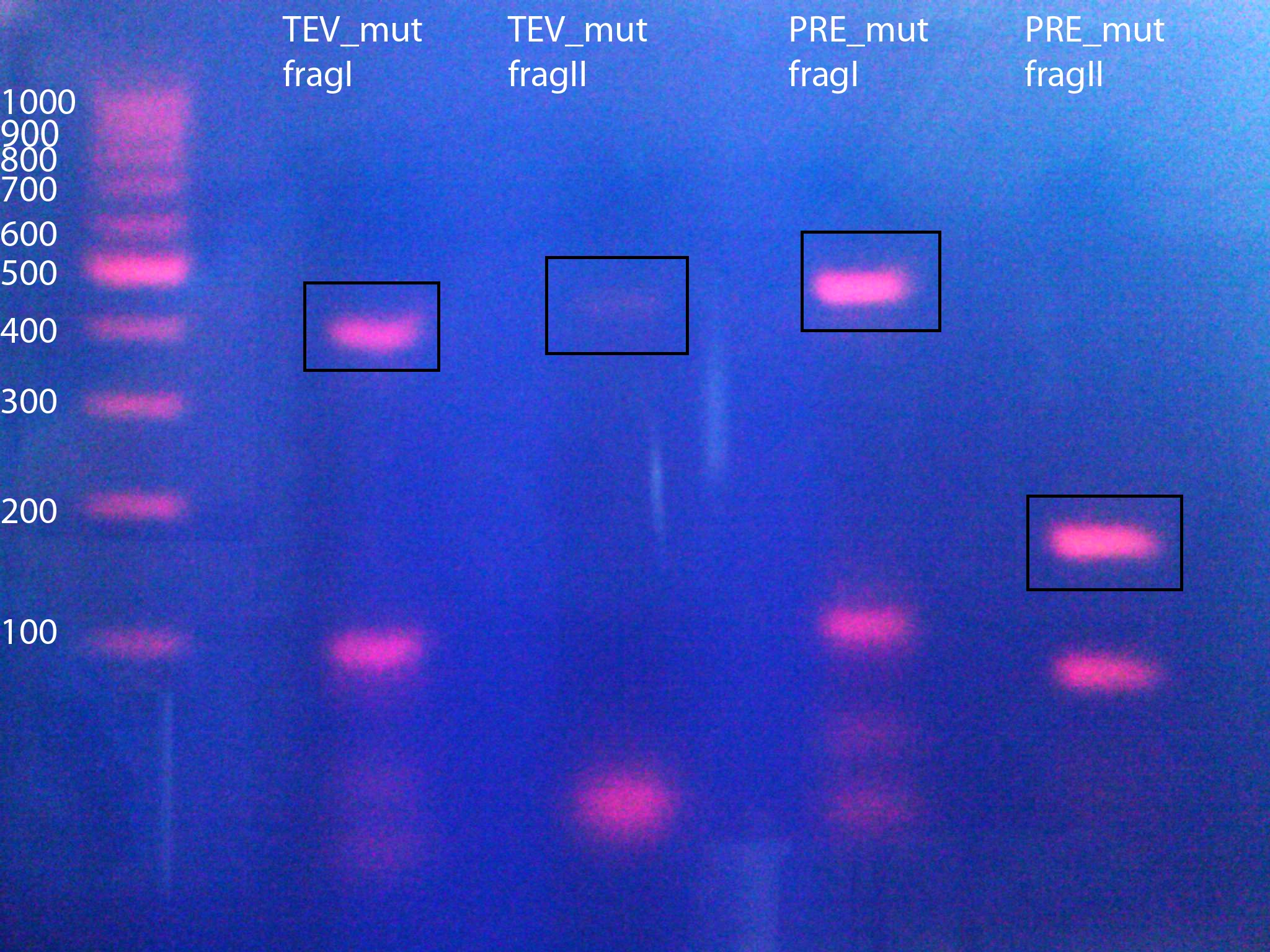
Mutagenesis of 14_3C and TEV proteases to remove iGEM restriction sites from the proteases and introduction of iGEM restriction sites
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim:
- Removal of iGEM restriction sites from proteases, amplifying protease fragments with iGEM restriction sites
Materials:
- Plasmid 1: pET9d_Thrombin-CS_XbaI-TEV-Protease_BamHI
- Primers: (1) f_TEV_ACCAGC , r_TEV_iGEM_BahmHI (2) r_TEV_ACCAGC , f_TEV_AraFusion_NgoMIV
- Plasmid 2: pGEX-3_14_3C
- Primers: (1) f_14_3C_ACCAGC, r_14_3C_iGEM_BamHI (2) r_14_3C_ACCAGC, f_14_3C_AraFusion_NgoMIV
Used method:
PCR
- Template: 1µl = 3,6 ng
- Nucleotides: 1µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 32,5µl of pure water
- 0,5µl TaqPol
Program: iGEM001
- Denat: 3min 94°C
- 5x:
Denat: 45sec 94°C
Anneal:45sec 53°C
Extend:45sec 72°C
- 25x:
Denat: 45sec 60°C
Anneal:45sec 60°C
Extend:45sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products (10µl) on 2% agarose gel
Expected Fragments:
TEV:
- TEV_mut_fragI: 360bp
- TEV_mut_fragII: 400bp
14_3C:
- 14_3C_mut_fragI: 420bp
- 14_3C_mut_fragII: 153bp
Further going:
- 40µl of PCR products left:*20µl for preparative agarose gel (2%)
- 20µl for PCR purification Kit
- Assembly PCR of purificated products to produce NgoMIV_iGEM_TEV-Protease_iGEM_BamHI and NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI
Amplification of Arabinose Induction System from pBAD_iGEMexpress
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Aim:
- Amplificarion of an arabinose induction system (AraC) from pBAD_iGEMexpress plasmid, produces a 1273bp fragment
Materials:
- Plasmid: pBAD_iGEMexpress (Nr.4)
- Primers: f_AraC_HindIII_iGEM , r_AraC_NgoMIV
Used method:
PCR
- Template: 1µl = 7,2 ng
- Nucleotides: 1 µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 32,5µl of pure water
- 0,5µl TaqPol
Program: iGEM002
- Denat: 3min 94°C
- 5x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
- 25x:
Denat: 60sec 60°C
Anneal:60sec 60°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products (10µl) on 1% agarose gel
Expected Fragments: HindIII_iGEM_AraC_NgoMIV 1273bp
Further tasks:
- 30µl of PCR product left: (1) Preparative Agarose Gel + Extraction(2) Digestion of fragment with HindIII and NgoMIV and gel purification (3) Ligation with (digested) NgoMIV_14_3C-Protease_iGEM_BamHI or NgoMIV_TEV_iGEM_BamHI fragments (see entry above).
EDIT:
The HindIII_iGEM_AraC_NgoMIV fragment was purificated from a preparative gel (1.5%)
Concentration:12,5ng/µl
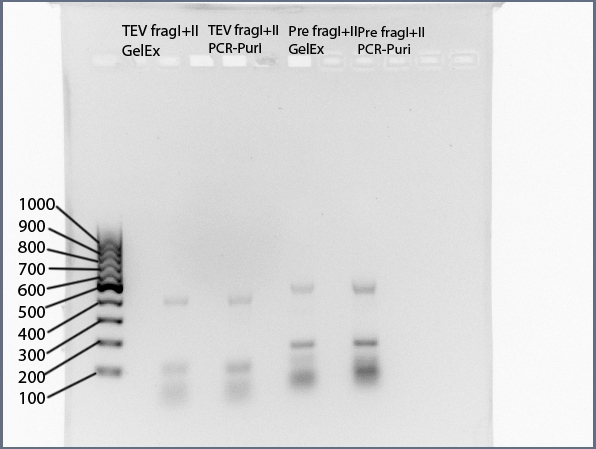
Annealing of TEV_mut_fragI and TEV_mut_fragII /14_3C_mut_fragI and 14_3C_mut_fragII
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Materials:
TEV:
- TEV_mut_fragI: 360bp (40µl)
- TEV_mut_fragII: 400bp (40µl)
Primers for PCR:
- f_TEV_AraFusion_NgoMIV
- r_TEV_iGEM_BamHI
HRV 14_3C:
- 14_3C_mut_fragI: 420bp (40µl)
- 14_3C_mut_fragII: 153bp (40µl)
Primers for PCR:
- f_14_3C_AraFusion_NgoMIV
- r_14_3C_iGEM_BamHI
Used method:
1. 20µl of EACH fragment were purificated using a preparative agarose gel followed by extraction with "NucleoSpin ExtractII"-KIT and the other 20µl of EACH fragment were purificated with PCR-purification unsing "NucleoSpin ExtractII"-KIT.
2. Annealing of purificated primers using PCR (program iGEM002):
- Template: Everything from purification (~15µl) = 4 reaction batches: 2x for TEV fragments, 2x 14_3C fragments from each purification method, respectively.
- Nucleotides: 1 µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 17,5µl of pure water
- 0,5µl TaqPol
Program: iGEM0002
- Denat: 3min 94°C
- 5x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
- 25x:
Denat: 60sec 60°C
Anneal:60sec 60°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products (5µl) on 1,5% analytical agarose gel
Expected Fragments
for TEV:
NgoMIV_iGEM_TEV-Protease_iGEM_BamHI - 760bp
for 14_3C:
NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI - 573bp
Summary:
Ligation of TEV_mut_fragI and TEV_mut_fragII did NOT work.
Ligation of 14_3C_mut_fragI and 14_3C_mut_fragII did work.
The two fractions of ligated 14_3C fragments were combined and PCR-purificated using "NucleoSpin ExtractII"-KIT (Concentration: 25ng/µl)
Further tasks:
Digestion of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI fragment with NgoMIV and BamHI
Starting a new PCR for to produce new TEV-mut_fragI and TEV_mut_fragII fragments!!
Planning and accomplishing the PCRs of pBAD-mYFP Venus with Arabinosis and pEX_HisII with Lac
Investigators: Nicole, Nadja
Aim:To get Biobricks with Arabinosis and IPTG induction
Time: 2011-08-01,14:00-19:00
Materials:
- Primer: 1. pr_Ara_Xba1, pf_Ara_EcoR1 and 2. pf_IPTG_EcoR1, pf_IPTG_Xba1
- Plasmids: 1. pBAD-mYFP Venus and 2. pEX_HisII
Method:PCR
1.pBAD-mYFP Venus with Ara
- 1,25 µl pBAD-mYFP Venus (1:10, 10,8 ng/µl)
- 1,00 µl dNTPs
- 5,00 µl Buffer
- 2,50 µl pr_Ara_XbaI
- 2,50 µl pf_Ara_EcoRI
- 2,00 µl MgCl2
- 0,50 µl Polymerase S
- 35,25 µl H2O
2. pEX_HisII with Lac
- 1,00 µl pEX_HisII(1:20, 9,6 ng/µl)
- 1,00 µl dNTPs
- 5,00 µl Buffer
- 2,50 µl pf_IPTG_EcoRI
- 2,50 µl pf_IPTG_XbaI
- 2,00 µl MgCl2
- 0,50 µl Polymerase S
- 35,50 µl H2O
3. Program for both: IGBIOB1 (30 cycles)
| Step | Temperature | Time |
|---|---|---|
| Hot start | 94°C | hold |
| Initial Denaturation | 94°C | 3min (180s) |
| Denaturation | 94°C | 20s |
| Annealing | 44°C | 40s |
| Extension | 72°C | 73s |
| Final Extension | 72°C | 600s |
Further tasks:
- See PCR results on agarose gel and do a gel extraction
51th Labday 2011-08-02
Transformation of BBa_I763007 in E. coli XL1-Blue
Investigators: Jessica, Steffi, Vanessa
Aim:Transformation of plasmid BBa_I763007 (containg Lambda promoter, RBS and RFP) in E. coli XL1-Blue cells to produce glycerol stocks for further use
Time: 2011-08-02,
Materials:
Method:
Further tasks:
- Picking clones for overnight culturing
- Producing glycerol stocks
Repetition of the PCRs of pBAD-mYFP Venus with Arabinosis and pEX_HisII with Lac because of less yield further planning and accomplishing the PCR of BBa_I763007 as it is a constitutive one
Investigators: Nadja, Nicole
Aim:To increase the yield and to get a constitutive Biobrick
Time:2011-08-02
Materials:
- Primer: 1. pr_Ara_Xba1, pf_Ara_EcoR1 and 2. pf_IPTG_EcoR1, pf_IPTG_Xba1 and 3. Pr_constitutive_XbaI, pf_constitutive_EcoRI
- Plasmids: 1. pBAD-mYFP Venus and 2. pEX_HisII and 3. BBa_1763007
Method:PCR
1. pBAD-mYFP Venus with Ara
- 1,25 µl pBad (1:10, 10,8ng/µl)
- 1,00 µl dNTPs
- 5,00 µl Buffer
- 2,50 µl pr_Ara_Xba1
- 2,50 µl pf_Ara_EcoR1
- 2,00 µl MgCl2
- 0,50 µl Polymerase S
- 35,25µl H2O
2. pEX_HisII with Lac
- 1,00 µl pEX (1:20, 9,6 ng/µl)
- 1,00 µl dNTPs
- 5,00 µl Buffer
- 2,50 µl pf_IPTG_EcoR1
- 2,50 µl pf_IPTG_Xba1
- 2,00 µl MgCl2
- 0,50 µl Polymerase S
- 35,50 µl H2O
1. BBa_1763007 -constititive
- 1,00 µl BBa_1763007
- 1,00 µl dNTPs
- 5,00 µl Buffer
- 2,50 µl pr_constitutive_XbaI
- 2,50 µl pf_ constitutive_EcoR1
- 2,00 µl MgCl2
- 0,50 µl Polymerase S
- 35,5 µl H2O
3. Program for 1. pBAD-mYFP Venus and 2. pEX_HisII: IGBIOB1 (30 cycles)
| Step | Temperature | Time |
|---|---|---|
| Hot start | 94°C | hold |
| Initial Denaturation | 94°C | 3min (180s) |
| Denaturation | 94°C | 20s |
| Annealing | 44°C | 40s |
| Extension | 72°C | 73s |
| Final Extension | 72°C | 600s |
4. Program for BBa_1763007 -constititive: IGBIOC1 (30 cycles: 2-step PCR, first run10x, second run 20x)
| Step | Temperature | Time |
|---|---|---|
| Hot start | 94°C | hold |
| Initial Denaturation | 94°C | 3min (180s) |
| Denaturation | 94°C | 20s |
| Annealing | 43°C | 40s |
| Extension | 72°C | 45s |
| Denaturation | 94°C | 20s |
| Annealing | 54°C | 40s |
| Extension | 72°C | 45s |
| Final Extension | 72°C | 600s |
Further tasks:
- See PCR results on agarose gel and do a gel extraction
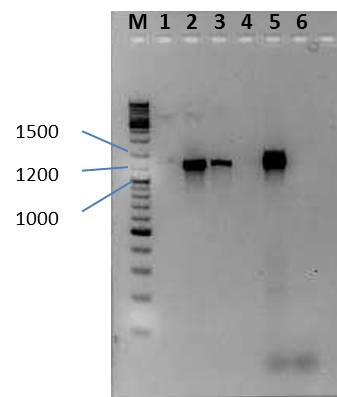
Agarose gel electrophoresis:
Gel 1
- 5 µl PCR + 1 µl 6x loading dye
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 10 | |
| 1 | - | - | - |
| 2 | PCR Arabinose promotor | 6 | ~1200 |
| 3 | PCR Arabinose promotor | 6 | ~1200 |
| 4 | - | - | - |
| 5 | PCR Lac promotor | 6 | ~1400 |
| 6 | PCR Lac promotor | 6 | ~1400 |
Gel 2
- 5 µl PCR + 1 µl 6x loading dye
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| 1 | PCR Lambda promotor from BBa_I763007 | - | ? |
| 2 | PCR Lambda promotor from BBa_I763007 | - | ? |
| 3 | - | - | |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 10 |
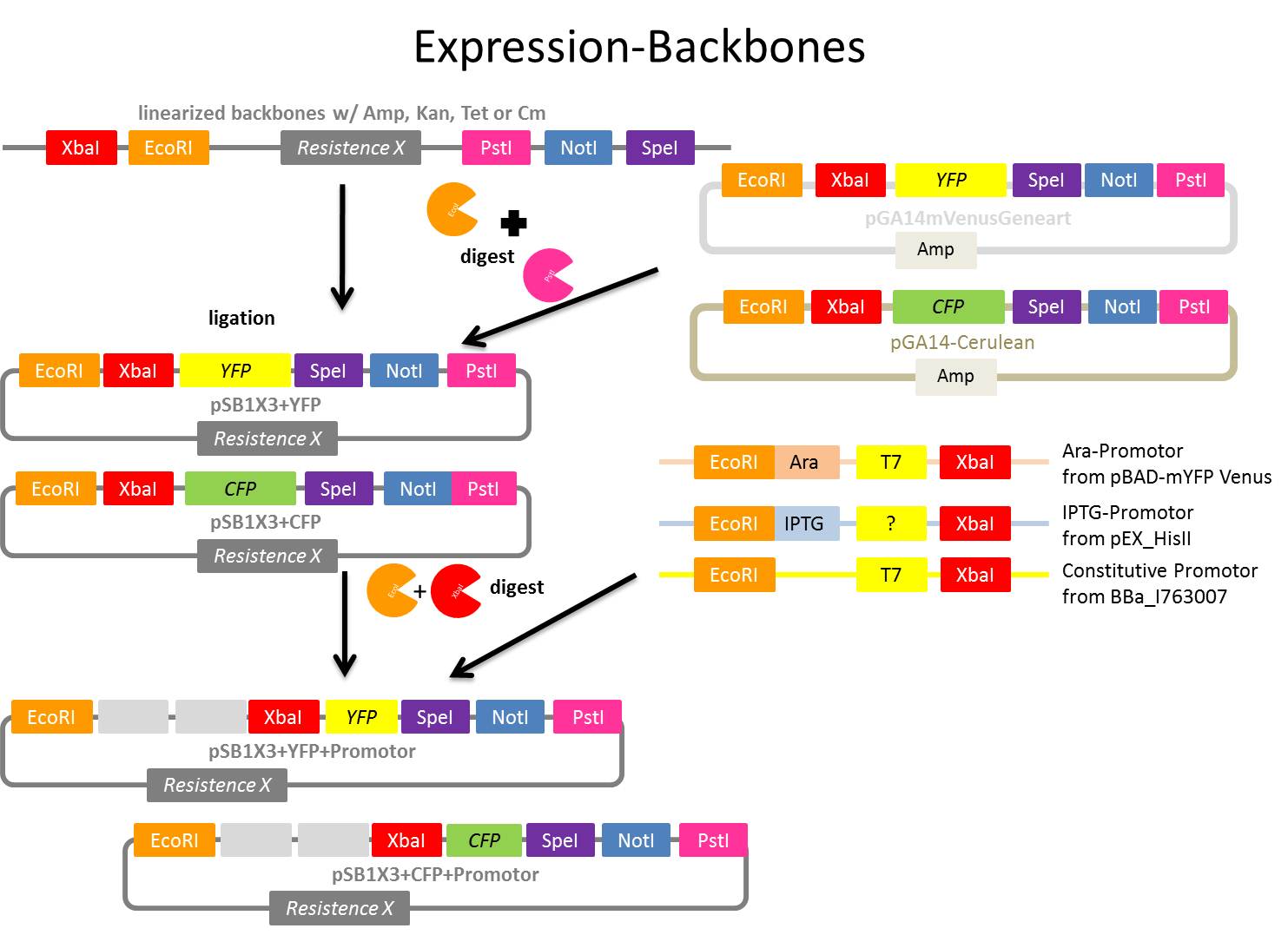
Preparing of linearized backbones (pSB1A3, pSB1K3 and pSB1T3) to produce vectors for further applications
Investigators: Nadja, Nicole
Time: 2011-08-02,
Aim: Using linearized backbones for further transformations, iGEM restrictions sites are necessary
Idea:
Producing vectors with different antibiotic resistances (ampicillin, kanamycin and tetracyclin) and iGEM restriction sites to
1. have the choice which resistance you want and then the possibility to clone each gene of interest easily in the chosen vector
2. to clone inducible and constitutive promoter systems in it and use this as expression vector for further experiments
Steps:
1. Restriction enzyme digestion
2. Ligation with reporter gene
3. Transformation and miniprep afterwards
Materials:
- Linearized backbones from iGEM Registry of Standard biological parts (part of Spring 2010 DNA distribution kits) - only with EcoRI and PstI restriction sites
- pSB1A3 - ampicillin resistance
- pSB1T3 - tetracycline resistance
- pSB1K3 - kanamycin resistance
- pSB1C3 - we already got from KUK lab
- Manual ‘Spring 2011 DNA distribution’, see: http://partsregistry.org/Help:Spring_2011_DNA_distribution or File:UP Spring 2011 DNA distribution.pdf
Restriction enzyme digestion of linearized plasmid backbones (pSB1A3, pSB1K3 and pSB1T3)
Investigators: Nadja, Nicole
Time: 2011-08-02,
Aim:Producing vectors, which carry tetracycline, kanamycin and ampicillin resistance genes and have all iGEM restriction sites
Materials:
- Linearized backbones from iGEM Registry of Standard biological parts (part of Spring 2010 DNA distribution kits) à only with EcoRI and PstI restriction sites
- pSB1A3, pSB1T3, pSB1K3
- Manual ‘Spring 2011 DNA distribution’, see: http://partsregistry.org/Help:Spring_2011_DNA_distribution or File:UP Spring 2011 DNA distribution.pdf
- NEB Buffer 2
- Purified BSA (NEB)
- EcoRI (NEB)
- pstI (NEB)
- dH2O
Method:
1. Enzyme master mix
- 5 µl NEB Buffer 2
- 0.5 µl BSA
- 0.5 µl of each EcoRI, PstI
- 18.5 µl dH2O
- mix 4 µl linearized plasmid backbone (25 ng/ µl) with 4 µl enzyme master mix
2. Reaction conditions
- 30 min at 37°C by 750 rpm
- 20 min at 80°C by 750 rpm
Further tasks:
- Gel electrophoresis
- DNA extraction
- Ligation with reporter genes CFP and YFP
- Transformation
- Miniprep
- Sequencing
Gel electrophoresis of digested (linearized) plasmid backbones (pSB1A3, pSB1K3 and pSB1T3)
Investigators: Nadja, Nicole
Time: 2011-08-02,
Aim:Producing vectors, which carry tetracycline, kanamycin and ampicillin resistance genes and have all iGEM restriction sites, through cloning CFP resp. YFP in the linearized plasmid backbones.
Materials:
- linearized plasmids backbones (pSB1A3, pSB1K3, pSB1T3) digested with EcoRI and PstI
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder GeneRuler, 100bp plus (1:10) (Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of one 1 % and one 1.5 % agarose gels
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- 1.5 % gel: 0.75 g in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
2. Loading gels and running
- Add 6 µl Loading dye to each 30 µl sample
- Gene Ruler DNA ladder
- Running conditions: 100 V, approx. 45 min
3. Loading of gels
| gel 1 (1 %) | gel 2 (1.5 %) | ||||||
|---|---|---|---|---|---|---|---|
| lane | Sample | Volume in µl | Expected size in bp | Sample | Volume in µl | Expected size in bp | |
| 1 | marker | marker | 6 | ||||
| 2 | - | - | - | - | - | - | |
| 3 | pSB1T3 | 36 | 2206 | CFP | 36 | ||
| 4 | - | - | - | - | - | - | |
| 5 | pSB1A3 | 36 | 1862 | YFP | 36 | ||
| 6 | - | - | - | - | - | - | |
| 7 | pSB1K3 | 36 | 1978 | - | - | - | |
Results:
[[File:]] [[File:]]
The bands were excised and purified using the NucleoSpin Extract II (Macherey-Nagel) extraction Kit.
Further Tasks:
- Ligation of CFP resp. YFP in each linearized backbone
- Transformation
- Minprep and production of glycerol stocks
52th Labday 2011-08-03
Transformation of generated pPDV089 (strategy 2)
Investigators: Leif
Aim:amplification of pPDV089
Time: 13:30-15:00
Method:
- addition of 10 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones
Ligation of pBAD-mYFP Venus and pEX_HisII with pSB1_K3 and pSB1_A3
Investigators:Nadja, Nicole
Aim:Build Biobricks inducible with IPTG or Arabinosis and detectible with YFP or CFP
Time:2011-08-02,10:00-13:00
Materials:
- T4 ligase
- 10x ligase buffer
- vectors: pSB1_K3 and pSB1_A3
- insert: pBAD-mYFP Venus and pEX_HisII
Method:
- total volume of 20µl
- 1µl T4 ligase
- 2µl 10x ligase buffer
- 5µl vector
- 3µl insert
- fill up to 20µl with H20
- incubation over night at 18°C
Further tasks:
- over night culture
- miniprep
- sequenzing
53th Labday 2011-08-04
digestion of vector pARW089 (strategy 2)
Investigators: Leif
Aim: digestion of pARW089
Time: 2011-08-04,10:00-12:00
Digestion of vector pARW089 with NarI (EheI isoschizomere) and AatII
- 20 µl sample
- NEB 10x buffer (2 µl)
- Buffer M 10x buffer (2 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 13 µl H2O
- 2 h at 37°C, then over night in the fridge
Wrong buffer, so the experiment was rerun.
digestion of PCR products vector pARW089 (strategy 2)
Investigators: Leif
Aim: digestion of pARW089
Time: 2011-08-04,10:00-12:00
Digestion of vector pARW089 with NarI (EheI isoschizomere) and AatII
- 20 µl sample
- NEB 10x buffer (2 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 13 µl H2O
- 2 h at 37°C, then over night in the fridge
Further Tasks:
- gel electrophoresis and purification of the two digested fragments
- Problem: Shaker was on, so no digest possible!
Investigators: Stefan
Aim:digestion of pARW089
Time: 2011-08-04,10:00-12:00
Digestion of vector pARW089 with NarI (EheI isoschizomere) and AatII
- 40 µl sample
- NEB 4 10x buffer (4 µl)
- 1 µl restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 32 µl H2O
- 2 µl pARW089 (undiluted sample)
2 h at 37°C, then heat over night in the fridge
Further Tasks:
- gel electrophoresis and purification of the two digested fragments
54th Labday 2011-08-05
Digestion of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV fragments and pJC354-NheI-143C-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul, Sebastian
Materials:
NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI - 573bp
HindIII_iGEM_AraC_NgoMIV - 1273bp
pJC354-NheI-143C-Xho_blaFL_GGH5 vector (contains 14_3C cleavage site)
Digestion protocol:
1: NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (25ng/µl):
- 30µl NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 12.5µl pure water
- =50µl
2: HindIII_iGEM_AraC_NgoMIV (12.5ng/µl)
- 40µl HindIII_iGEM_AraC_NgoMIV
- 1µl HindIII
- 1µl NgoMIV
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 1.5µl pure water
- =50µl
3: pJC354-NheI-143C-Xho_blaFL_GGH5 vector (270ng/µl)
- 4µl pJC354-NheI-143C-Xho_blaFL_GGH5 vector (contains 14_3C cleavage site)
- 1µl HindIII
- 1µl BamHI
- 5µl 10x buffer
- 0.5µl 100x BSA
- 38.5µl pure water
- =50µl
-->The reaction was allowed to proceed for 2h!
Result:
Resolving of digested fragments (50µl) and digested vector (50µl) on 1.5% and 1% preparative agarose gels, respectively.
1: NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (band 5) and HindIII_iGEM_AraC_NgoMIV (band 3) fragments
2: pJC354-NheI-143C-Xho_blaFL_GGH5 vector
Summary:
The bands corresponding to digested fragments were excissed and purificated with PCR-purification unsing "NucleoSpin ExtractII"-KIT.
Further tasks:
Triple ligation of digested fragments
digest pSB1A3, pSB1K3 (clone 1-16)
Investigators: Niels
Aim: prove of Insert
Digestion protocol: 16x
- 5µl DNA - pSB1A3,pSB1K3
- 0,5µl XbaI
- 0,5µl EcoRI
- 2µl 10x buffer = NEB 4
- 12.5µl pure water
- total: 20µl
37°C for 1h
Result:
Further tasks: sequencing
Restriction enzyme digestion pARW089 for Phage Display strategy II
Investigators: Leif
Time: 2011-08-05,10:00-12:30
Aim: Restriction enzyme digestion of pARW089 according to the protocol by Nadine from the 2011-07-22
Materials:
- DNA: 5 µL
- NEB Buffer 4 (10x): 4 µL
- Enzyme AatII: 0.8 µL
- Enzyme NarI: 2 µL
- H2O: 28.2 µL
Total Volume: 40 µL
Results: Incubation of the digestion for 2 h with 750 rpm caused a breakdown of the restriction enzymes. The expreriment has to be repeated.
Miniprep of overnight cultures from ligation of pARW089 + mutated mdnA and test digest
Investigators: Jessica
Time: 2011-08-05, 10:00-18:30
Aim: DNA for sequencing and confirmation of insert
1. Miniprep:
- 20 overnight cultures (1A-2a(2 h digest, clone a),1A-2b,2A-2a,...,5A-3b)
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| 1A-2a | 435.9 |
| 2A-2a | 480.1 |
| 3A-2a | 464.3 |
| 4A-2a | 362.7 |
| 5A-2a | 433.9 |
| 1A-3a | 528.3 |
| 2A-3a | 489.5 |
| 3A-3a | 487.3 |
| 4A-3a | 422.7 |
| 5A-3a | 537.3 |
| 1A-2b | 422 |
| 2A-2b | 297.4 |
| 3A-2b | 538.2 |
| 4A-2b | 427 |
| 5A-2b | 452 |
| 1A-3b | 375.5 |
| 2A-3b | 549.9 |
| 3A-3b | 307.8 |
| 4A-3b | 450.1 |
| 5A-3b | 462.3 |
2. Preparation of glycerol stocks:
- adding 300 µl glycerol to 700 µl culture
3. Digest:
- 2µl DNA (10 samples, only clone a)
- 0,5µl NarI
- 0,5µl AatII
- 2µl 10x buffer NEB 4
- 15µl H2O
- total: 20µl
- 37°C for 1h
4. Agarose gel electrophoresis:
- 1% agarose gel
- 1 h at 115 V
Samples
- 20 µl digest + 4 µl 6x loading dye
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 10 | |
| 1 | 1A-2a | 24 | 10296, 116 |
| 2 | 2A-2a | 24 | 10296, 116 |
| 3 | 3A-2a | 24 | 10296, 116 |
| 4 | 4A-2a | 24 | 10296, 116 |
| 5 | 1A-3a | 24 | 10296, 116 |
| 6 | 5A-2a | 24 | 10296, 116 |
| 7 | 2A-3a | 24 | 10296, 116 |
| 8 | 3A-3a | 24 | 10296, 116 |
| 9 | 4A-3a | 24 | 10296, 116 |
| 10 | 5A-3a | 24 | 10296, 116 |
Result:
- Inserts could be confirmed for samples from clone a
Further tasks:
- sequencing to determine mutation rate
Miniprep of overnight cultures from ligation of pBAD-mYFP Venus and pEX_HisII with pSB1_K3 and pSB1_A3
Investigators: Nicole, Nadja
Time: 2011-08-05
Aim: DNA for sequencing and confirmation of insert
Materials/Methods:
1. Miniprep:
- 16 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
2. Preparation of glycerol stocks:
- adding 300 µl glycerol to 700 µl culture
Further tasks:
- sequenzing
55th Labday 2011-08-06
2nd Mutagenesis of TEV proteases to remove iGEM restriction sites from the proteases and introduction of iGEM restriction sites
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul
Aim:
- Removal of iGEM restriction sites from proteases, amplifying protease fragments with iGEM restriction sites
Materials:
- Plasmid 1: pET9d_Thrombin-CS_XbaI-TEV-Protease_BamHI
- Primers: (1) f_TEV_ACCAGC , r_TEV_iGEM_BahmHI (2) r_TEV_ACCAGC , f_TEV_AraFusion_NgoMIV
Used method:
PCR
- Template: 1µl = 3,6 ng
- Nucleotides: 1µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 32,5µl of pure water
- 0,5µl TaqPol
Program: iGEM001
- Denat: 3min 94°C
- 5x:
Denat: 45sec 94°C
Anneal:45sec 53°C
Extend:45sec 72°C
- 25x:
Denat: 45sec 94°C
Anneal:45sec 60°C
Extend:45sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products (50µl) on 2% preparative agarose gel
Expected Fragments:
TEV:
- TEV_mut_fragI: 360bp
- TEV_mut_fragII: 400bp
Further going:
- gel extraction with NucleoSpin Extract II
- Assembly PCR of purificated products to produce NgoMIV_iGEM_TEV-Protease_iGEM_BamHI
2nd Annealing of TEV_mut_fragI and TEV_mut_fragII
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Paul
Materials:
TEV:
- TEV_mut_fragI: 360bp (40µl)
- TEV_mut_fragII: 400bp (40µl)
Primers for PCR:
- f_TEV_AraFusion_NgoMIV
- r_TEV_iGEM_BamHI
Used method:
1. 50µl of EACH fragment were purificated using a preparative agarose gel followed by extraction with "NucleoSpin ExtractII"-KIT.
2. Annealing of purificated primers using PCR (program iGEMMED):
- Template: 5µl TEV_mut_fragI and 10µl TEV_mut_fragII = 1 reaction batch.
- Nucleotides: 1 µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 18,5µl of pure water
- 0,5µl TaqPol
Program: iGEMMED
- Denat: 3min 94°C
- 3x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
- 28x:
Denat: 60sec 94°C
Anneal:60sec 65°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Result:
Resolving of PCR products (50µl) on 1,5% preparative agarose gel
Expected Fragments
for TEV:
NgoMIV_iGEM_TEV-Protease_iGEM_BamHI - 760bp
GelDoc documentation was not possible. 3 bands were visible:
1 band (1) at 700 - 800 bp (NgoMIV_iGEM_TEV-Protease_iGEM_BamHI),
1 band (2) at 500 - 600 bp (unexpected,)
1 band (3) at 300 - 400 bp (TEV_mut_fragI and TEV_mut_fragII)
Summary:
Ligation of TEV_mut_fragI and TEV_mut_fragII did work.
Unexpected band at 500 - 600 bp.
Further tasks:
Digestion of NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment and of the unexpected band with NgoMIV and BamHI.
Digestion of NgoMIV_iGEM_TEV-Protease_iGEM_BamHI and of the unexpected band from the 2nd annealing TEV PCR
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Sascha
Materials:
NgoMIV_iGEM_TEV-Protease_iGEM_BamHI - 760bp
unexpected band - 500-600 bp
Digestion protocol:
- 30µl NgoMIV_iGEM_TEV-Protease_iGEM_BamHI / 30µl unexpected band
- 1µl NgoMIV
- 1µl BamHI
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 12.5µl pure water
- =50µl
- 5µl vector
-->The reaction was allowed to proceed for 2h and was resolving (50µl) on 1,5% preparative agarose gel.
Result:
No visible bands on 1.5% preparative agarose gels.
Further tasks:
New PCR for to produce new TEV-mut_fragI and TEV_mut_fragII fragments!!
Repeated PCR of mdnA and gene III for phage display (strategy 2)
Investigator: Sabine, Sandrina
Time: 2011-08-06,12:00-14:00
Aim:
- amplification of mdnA with NarI and AgeI restriction sites (strategy 2)
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (mdnaA, strategy 2)
- primer: pf_geneIII_NgoMIV and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl Vector pARW089/Vector 100blaKDIR
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
Further tasks:
- purification
- digestion
digestion of PCR products (strategy 2)
Investigator: Sabine, Sandrina
Aim:
digestion of mdnA and gene III for getting an mdnA-geneIII fusion gene with rfc25 restrition sites after ligation
Time: 2011-08-06,14:00-15:30
digestion enzymes:
- digestion of mdnA (strategy 2) with NarI and AgeI
- digestion of geneIII (strategy 2) with NgoMIV and AatII
reaction components:
- 4 µl NEB 10x buffer
- 1 µl per restriction enzyme
- 30 µl PCR product
- 4 µl H²O
reaction conditions:
- 1 h for PCR fragments
- 37°C NarI, AgeI, NgoMIV and AatII digestion
Further Tasks:
- gel electrophoresis for purification of the digested fragments and vectors
gel electrophoresis of digested fragments and digested pARW089
Investigator: Sandrina
Aim:
- control and purification of digested PCR fragments
- control and purification of digested pARW089 (2011-08-05)
Time: 2011-08-06,14:00-18:00
Results:
- mdnA (NarI and AgeI, stategy 2): ca 200 bp, but estimation difficult, because no GelDoc available (weekend)
- geneIII (NgoMIV and AatII): no band
- pARW089 : ca 10 kb, but estimation difficult, because no GelDoc available (weekend)
Further Tasks:
- repeat PCR of geneIII
- ligation
prepare samples for sequencing (pARW089 + mutated mdnA)
Investigators: Niels
Aim: mdnA_1 sequencing by eurofins mwg|operon
guidelines (eurofins mwg|operon)
Primer :2 pmol/µl (10 µl each sample)
- 2,4 µl (100 µM) sf_mdnA_1
- 117,6 µl water
Sample: 70 ng/µl (15 µl total)
- sample (cDNA ng/µl) - DNA µl ( ad water 15 µl)
- 1A-2a (435,9) - 2,4
- 2A-2a (480,1) - 2,2
- 3A-2a (464,3) - 2,3
- 4A-2a (362,7) - 2,9
- 5A-2a (433,9) - 2,4
- 1A-3a (528,3) - 1,99
- 2A-3a (489,5) - 2,15
- 3A-3a (487,3) - 2,15
- 4A-3a (422,7) - 2,48
- 5A-3a (537,3) - 1,95
Legend:
- example: 1A-2a
- 1A-2a
- sample 1-5 : different Mn-concentration (error-pone PCR)
- 1A-2a
- sample A-C : different error-pone PCR- appendage
- 1A-2a
- sample 2 or 3: 2h(or 3h) digest of pARW089 with AatII / NarI
- 1A-2a
- sample a or b : 1. clone(a) or 2. clone(b) from the same plate with: E. coli XL1 blue transformed with ligation of vector pARW089 and insert (error pone - PCR )
Further tasks: sequencing
56th Labday 2011-08-07
PCR purification of TEV_mut_fragI und II
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Stefan
Aim:
- PCR purification
Materials:
- Promega PCR clean-Up System
- TEV_mut_fragI (360bp), TEV_mut_fragII (400bp)
Used method:
Further going:
gel electrophoresisTEV_mut_fragI and II
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Stefan
Aim:
- TEV_mut_fragI (360bp), TEV_mut_fragII (400bp)
Materials:
- use small raq at 80V
Used method:
- no picture could be taken due to the weak UV signal
Further going:
Annealing PCR TEV_mut_fragI and II
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Stefan
Aim:
- TEV_mut_fragI (360bp), TEV_mut_fragII (400bp)
Materials:
1. 50µl of EACH fragment were purificated using a preparative agarose gel followed by extraction with "NucleoSpin ExtractII"-KIT.
2. Annealing of purificated primers using PCR (program iGEMMED):
- Template: 5µl TEV_mut_fragI and 10µl TEV_mut_fragII = 1 reaction batch.
- Nucleotides: 1 µl of 10mM ready to use dNTP mix
- 5µl 10x Amplification buffer S
- 5µl 25mM MgCl2
- 2,5µl primers = 25pmol absolute (2,5µl of each primer = 5µl per tube)
- 18,5µl of pure water
- 0,5µl TaqPol
Program: iGEMMED
- Denat: 3min 94°C
- 3x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
- 28x:
Denat: 60sec 94°C
Anneal:60sec 65°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Used method:
Further going:
Ligation of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV and pJC354-NheI-143C-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Stefan
Aim:Triple-ligation of
NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (573bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-143C-Xho_blaFL_GGH5 vector
Materials:
- 3 µL 14_3C fragment
- 3 µL AraC fragment
- 1 µL pJC354
- 2 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- 10 µL H20
Used method:
ligation at room temperatur for 3h
Results:
band was very blurry on the gel, probably of contaminated running buffer
Further going:
Transformation of Ligation of pJC AraC and 14_3C in E. coli XL1 blue
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Stefan
Aim:transform the ligation into E. coli XL1 blue
Materials:
protocol:
addition of 10 µl ligation reaction to cells (XL1-blue, tet-resistance) in 1.5 ml Eppi,
incubation 25 min on ice,
heat shock 45 sec at 42°C,
incubation 2 min on ice,
addition of 750 µl LB medium,
incubation at 37 °C for 60 min in Eppendorf thermomixer at 750 rpm,
plating on LB medium with 1,5 % agar, 100 µg/ml chloramphenicol,
storage over night at 37°C
Result:
no colonies
Further going:
Repeated PCR gene III for phage display (strategy 2)
Investigator: Sabine
Time: 2011-08-07,11:00-13:00
Aim:
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_geneIII_NgoMIV and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl Vector pARW089 / pak100blaKDIR
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
- purification of PCR fragments with QIAquick Gel Extraction Kit (250)
Further tasks: digestion
digestion of PCR products and vector (strategy 2)
Investigator: Sabine
Aim:
- digestion of mdnA and gene III for getting an mdnA-geneIII fusion gene with rfc25 restrition sites after ligation
- digestion of pARW089 for ligation of mdnA/geneIII fusion gene into it (strategy 2)
Time: 2011-08-06,12:30-15:00
digestion enzymes:
- digestion of mdnA (strategy 2) with NarI and AgeI
- digestion of geneIII (strategy 2) with NgoMIV and AatII
- digestion of pARW089 (strategy 2) with NarI and AatII
reaction components:
- 5 µl NEB 10x buffer
- 1 µl per restriction enzyme
- 40 µl PCR product / 5 µl vector
- ad 50 µl water
reaction conditions:
- 1 h for PCR fragments
- 3 h for plasmids
- 37°C
Further Tasks:
- gel electrophoresis for purification of the digested fragments and vectors
prepare samples for sequencing (pSB1A3, pSB1K3)
Investigators: Niels
Aim: sequencing by eurofins mwg|operon
guidelines (eurofins mwg|operon)
Primer :2 pmol/µl (10 µl each sample)
Sample: 70 ng/µl (15 µl total)
- sample (cDNA ng/µl) - DNA µl ( ad water 15 µl)
- 1A3 CFP (204,5) - 5,13
- 1K3 CFP (216,2) - 4,86
- 1A3 YFP (235,7) - 4,45
- 1K3 YFP (213,6) - 4,92
Further tasks: sequencing
57th Labday 2011-08-08
test digest of pSB1K3
Time: 2011-08-08,11:00-16:00
Investigators: Vanessa, Steffi, Katharina, Nadine
Aim: prove of Insert (CFP or YFP)
Materials:
- pSB1K3 (clone 1 and 4 from last week ???)
- EcoRI, PstI, XbaI, HindIII, AatII
- Buffer 4
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Digestion protocol (XbaI, EcoRI)
- 5 µl DNA - pSB1K3 YFP or CFP (clone 4 or 1, respectively)
- 0.5 µl XbaI
- 0.5 µl EcoRI
- 2 µl 10x buffer = NEB 4
- 12.5 µl pure water
Digestion protocol (PstI, EcoRI)
- 5 µl DNA - pSB1K3+YFP or CFP (clone 4 or 1, respectively)
- 0.5 µl EcoRI
- 0.5 µl PstI
- 2 µl 10x buffer = NEB 4
- 12.5 µl pure water
Digestion protocol (AatII, HindIII)
- 5 µl DNA - pSB1K3 YFP or CFP (clone 4 or 1, respectively)
- 0.5 µl AatII
- 0.5 µl HindIII
- 2 µl 10x buffer = NEB 4
- 12.5 µl pure water
- total: 20µl
37°C for 3h
Production of one 1 %
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add 4 µl Loading dye to each 20 µl sample
- 15 µl DNA Ladder Mix
- Running conditions: 100 V, approx. 45 min
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | marker | ||
| 2 | pSB1K3+CFP (clone1), EcoRI, PstI | 24 | 812, 2163 |
| 3 | pSB1K3+CFP (clone1), XbaI, EcoRI | 24 | 7, 2968 |
| 4 | pSB1K3+CFP (clone1), AatII, HindIII | 24 | 721, 2254 |
| 2 | pSB1K3+YFP (clone4), EcoRI, PstI | 24 | 789, 2163 |
| 3 | pSB1K3+YFP (clone4), XbaI, EcoRI | 24 | 7, 2945 |
| 4 | pSB1K3+YFP (clone4), AatII, HindIII | 24 | 721, 2231 |
Result:
Further tasks: repeat the test digest tomorrow
sequencing pARW089 + mutated mdnA
Investigators: Niels,Steffi
Aim: sequencing by eurofins mwg|operon
samples prepared at 06.08.2011
Primer :2 pmol/µl 120 µl
sf_mdnA_1
pARW089 + mutated mdnA (15 µl total)
- sample ID : 2h
- 1A-2a - AKM001W020
- 2A-2a - AKM001W021
- 3A-2a - AKM001W022
- 4A-2a - AKM001W023
- 5A-2a - AKM001W024
- sample ID : 3h
- 1A-3a - AKM001W025
- 2A-3a - AKM001W026
- 3A-3a - AKM001W027
- 4A-3a - AKM001W028
- 5A-3a - AKM001W029
Results (arrived on 2011-08-11):
- no mutations could be found in mdnA
Assembly PCR of TEV_frag_I and TEV_frag_II
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sebastian, Stefan
Aim: Assembly PCR to mutate the EcoRI site
Materials:
- Primer:
- 2,5 µLf_TEV_AraFusion_NgoMIV (0.5 µmol stock)
- 2,5 µLr_TEV_iGEM_BamHI (0.5 µmol stock)
- 1 µL Fragment TEV I (approx. 2.5 ng)
- 1 µL Fragment TEV II (approx. 2.5 ng)
- 5 µL 10x polymerase buffer
- 5 µL 25 mM MgCl2
- 1 µL dNTP
- 0.5 µL T4 polymerase(Fermentas)
- 18.5 µL H20
Used method:
Program: iGEMMED
- Denat: 3min 94°C
- 3x:
Denat: 60sec 94°C
Anneal:60sec 53°C
Extend:60sec 72°C
- 28x:
Denat: 60sec 94°C
Anneal:60sec 65°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Further going:
analytical gel electrophoresis to confirm the correct size of bands
design primer for biobrick - mdnABC,mdnB,mdnC,mdnDE,mdnD,mdnE
Investigators: Katharina, Niels
Aim: design and order primer
Primer sequence
- pf_mdnABC_EcoRI_NotI_XbaI
GAATTCGCGGCCGCTTCTAGATGGCATATCCCAACGATC
- pf_mdnB_EcoRI_NotI_XbaI
GAATTCGCGGCCGCTTCTAGATGAAAGAATCGCCTAAAGTTG
- pf_mdnC_EcoRI_NotI_XbaI
GAATTCGCGGCCGCTTCTAGATGACCGTTTTAATTGTTAC
- pf_mdnE_EcoRI_NotI_XbaI
CAATCATCATATAACTCCGTAGATCTTCGCCGGCGCTTAAG
- pf_mdnD_EcoRI_NotI_XbaI
GTCAAAAAGGTCACGAAAGTAGATCTTCGCCGGCGCTTAAG
- pr_mdnABC_SpeI_NotI_PstI
GAAATCCTAGTTAACTCATAATACTAGTAGCGGCCGCTGCAG
- pr_mdnE_SpeI_NotI_PstI
CTGCAGCGGCCGCTACTAGTAGATATAAGAGTGGGTAAAATTC
- pr_mdnDE_SpeI_NotI_PstI
CTGCAGCGGCCGCTACTAGTATCAGCAAACCCTACTTAATTTC
- pr_mdnB_SpeI_NotI_PstI
GCGATCGCTGATTTTTTAGTTACTAGTAGCGGCCGCTGCAG
ordered my sigma
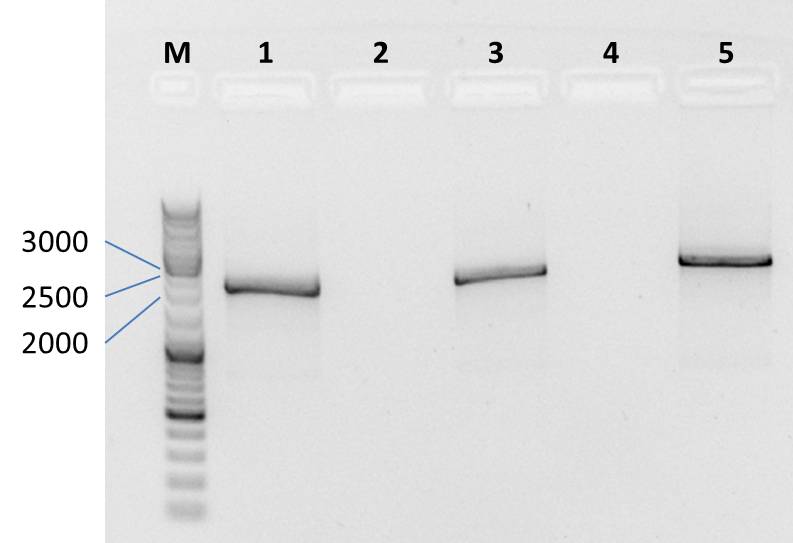
Ligation of mdnA/GeneIII-fusion gene into pARW089 for phage display and transformation of competent cells(strategy 2)
Investigators: Sandrina, Sabine
Aim:
- ligation of mdnA-geneIII fusiongene into pARW089 with digested fragments (see 2011-08-07)
- amplification of generated plasmids by transformation
Time: 10:00-18:00
Method:
- ligation-samples from 2011-08-07;
Protocol:
- 5 µl (ca 11 ng) digested vector pARW089 (NarI, AatII)
- 1 µl (ca 270 ng) fusion gene mdnA/geneIII
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 10 µl water
5 h at 16°C and 1 h at room temperature
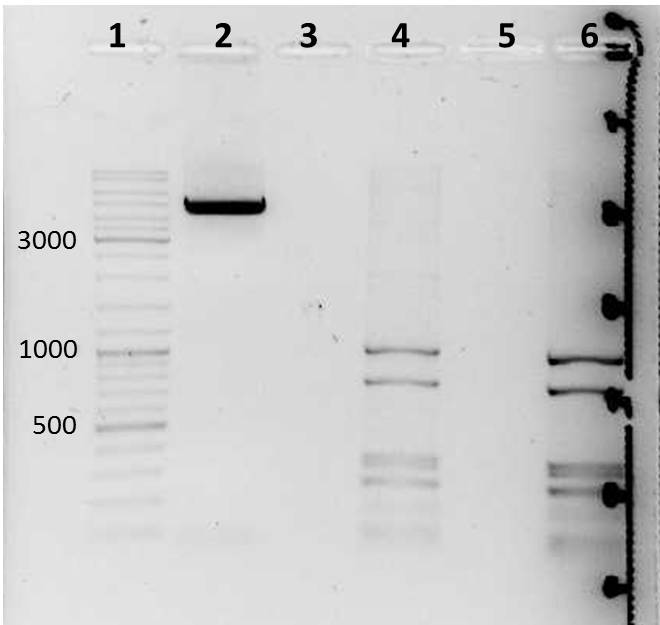
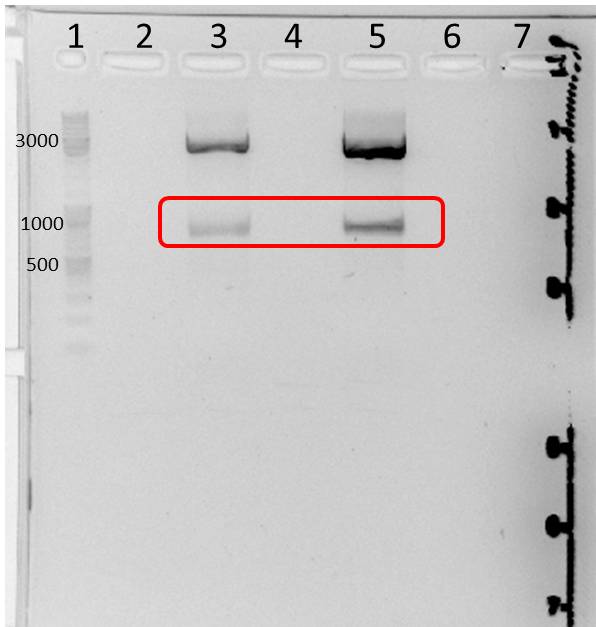
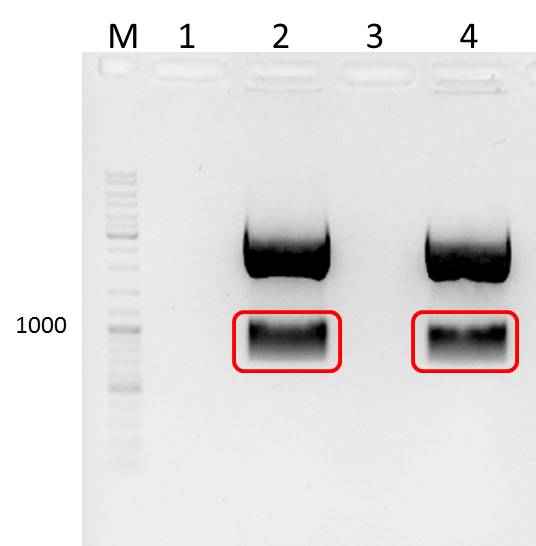
- two bands were observed after mdnA-geneIII ligation (red boxes)--> ligation with pARW089 was tried with both bands
transformation:
protocol:
addition of 10 µl ligation reaction to cells (XL1-blue, tet-resistance) in 1.5 ml Eppi,
incubation 25 min on ice,
heat shock 45 sec at 42°C,
incubation 2 min on ice,
addition of 750 µl LB medium,
incubation at 37 °C for 60 min in Eppendorf thermomixer at 750 rpm,
plating on LB medium with 1,5 % agar, 100 µg/ml ampicillin, 100 µg/µl tetracyclin
storage over night at 37°C
Further tasks:
test digestion
58th Labday 2011-08-09
test digest of pSB1K3+YFP or CFP and pSB1A3+YFP or CFP
Time: 2011-08-09,9:00-15:00
Investigators: Nadine, Vanessa, Steffi, Laura
Aim: prove of Insert (CFP or YFP)
Materials:
- pSB1K3 (clone 1 and 4 from last week ???), pSB1AK (clone 11 and 12 from last week ???)
- EcoRI, XbaI, HindIII, AatII, HaeII, BglI
- Buffer 4
- Buffer 2
- 100x BSA
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Digestion protocol (XbaI, EcoRI) (4x)
- 5 µl DNA - pSB1K3+YFP or CFP (clone 4 or 1, respectively) or pSB1A3+YFP or CFP (clone 12 or 11, respectively)
- 0.5 µl XbaI
- 0.5 µl EcoRI
- 2 µl 10x buffer = NEB 4
- 12.5 µl pure water
Digestion protocol (AatII, HindIII)
- 5 µl DNA - pSB1K3 YFP or CFP (clone 4 or 1, respectively) (2x)
- 0.5 µl AatII
- 0.5 µl HindIII
- 2 µl 10x buffer = NEB 4
- 12.5 µl pure water
- total: 20µl
Digestion protocol (HaeII, BglI) (4x)
- 5 µl DNA pSB1A3+YFP or CFP (clone 12 or 11, respectively)
- 0.5 µl HaeII
- 0.5 µl BglI
- 2 µl 10x buffer = NEB 2
- 0.2 µl 100xBSA
- 12.3 µl pure water
37°C for 2h
Production of one 1 % agarose gel
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add 4 µl Loading dye to each 20 µl sample
- 15 µl DNA Ladder Mix
- Running conditions: 100 V, approx. 45 min
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | marker | ||
| 2 | pSB1K3+CFP (clone1), not digested | 24 | |
| 3 | pSB1K3+CFP (clone1), XbaI, EcoRI | 24 | 7, 2968 |
| 4 | pSB1K3+CFP (clone1), AatII, HindIII | 24 | 721, 2254 |
| 5 | pSB1K3+YFP (clone4), XbaI, EcoRI | 24 | 7, 2945 |
| 6 | pSB1K3+YFP (clone4), AatII, HindIII | 24 | 721, 2231 |
| 7 | pSB1K3+YFP (clone4), not digested | 24 | |
| 8 | pSB1A3+CFP (clone11), not digested | 24 | |
| 9 | pSB1A3+CFP (clone11), XbaI, EcoRI | 24 | 15, 2911 |
| 10 | pSB1A3+CFP (clone11), HaeII, BglI | 24 | 1359, 1567 |
| 11 | pSB1A3+YFP (clone12), XbaI, EcoRI | 24 | 15, 2888 |
| 12 | pSB1A3+YFP (clone12), HaeII, BglI | 24 | 765, 924, 1188 |
Result:
Further tasks: repeat from beginning: w/ linearized backbones pSB1K3, pSB1A3 and pSB1T3
Gel purification of amplificated TEV protease
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sebastian
Aim: Clean up a pure fraction of TEV protease without iGEM-RS in the nucleotide sequence
Materials:
PCR and gel purification kit purchased by Promega (Wizard SV GEL and PCR Clean-Up System)
Used method:
Done as described in the manual
Results:
NgoMIV_TEV_iGEM_BamHI with a concentration of 6,9 ng/µl
Further going:
Amplification of TEV protease via PCR and digest with BamHI and NgoMIV
Ampilifcation of NgoMIV_TEV_iGEM_BamHI protease via PCR
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sebastian, Niels
Aim: Amplification of the TEV protease for digest with BamHI and NgoMIV
Materials:
1 µl Template - NgoMIV_TEV_iGEM_BamHI (6,9 ng/µl)
2,5 µl Primer 1 - f_TEV_AraFusion_NgoMIV (0,5 µM)
2,5 µl Primer 2 - r_TEV_iGEM_BamHI (0,5 µM)
5 µl 10x Reaction Buffer (Fermentas)
1 µl 10 mM dNTP's (Fermentas)
5 µl 25 µM MgCl2
0,5 µl DNA-Polymerase (Fermentas)
32,5 µl water
Used method:
Program: iGEM004
- Denat: 3min 94°C
- 30x:
Denat: 60sec 94°C
Anneal:60sec 65°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Further going:
PCR-Purification of the amplified DNA-Fragments and digest for ligation
PCR -Purification of amplified NgoMIV_TEV_iGEM_BamHI DNA-Fragements
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sebastian
Aim: Clean UP of the amplified fragments
Materials:
PCR-Clean up kit purchased by Promega(Wizard SV Gel and PCR-Up System)
Used method:
As described in the Manual of the kit
Results:
25 µl NgoMIV_TEV_iGEM_BamHI with 68,5 ng/µl
Further going: digest of the fragments and ligation for transformation
Ampilifcation of NgoMIV_TEV_iGEM_BamHI protease via PCR
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sebastian
Aim: 2nd amplification of the NgoMIV_TEV_iGEM_BamHI protease for digest with BamHI and NgoMIV
Materials:
1 µl Template - NgoMIV_TEV_iGEM_BamHI (6,9 ng/µl)
2,5 µl Primer 1 - f_TEV_AraFusion_NgoMIV (0,5 µM)
2,5 µl Primer 2 - r_TEV_iGEM_BamHI (0,5 µM)
5 µl 10x Reaction Buffer (Fermentas)
1 µl 10 mM dNTP's (Fermentas)
5 µl 25 µM MgCl2
0,5 µl DNA-Polymerase (Fermentas)
32,5 µl water
Used method:
Program: iGEM005
- Denat: 3min 94°C
- 30x:
Denat: 60sec 94°C
Anneal:60sec 63°C
Extend:60sec 72°C
- Final Extend: 10min 72°C
Further going:
PCR-Purification of the amplified DNA-Fragments and digest for ligation
ELISA to test the anti myc-tag antibodies 9E10
Investigators: Sebastian
Aim: Testing the reactivity of the 9E10 antibody fractions
Method:
Coating of an ELISA (96-well microtiter plate) with 50 µl/well 5 mg/ml FITC-BSA and incubation over night
Blocking of the free binding sites with 50 µl/well PBS-5% NKS 0,0025% phenolred for 1 hour
Incubation with different ScFv tagged with myc (unkown concentration)for 1 hour
- A1-H4 - anti FITC ScFv
- A5-H8 - Z6.1 (anti-FITC ScFV)
- A9-H12 - GST-tagged protein
Incubation with the tracer antibody 9E10 (different fractions in different lines (A-H) for 1 hour
Incubation with goat anti mice antibody labeled with HRP for 1 hour
Addition of the HRP-substrate (1 mg/ml OPD, 0,01 % H2O2, in 0,1 M Na-Citrate Buffer pH 5) for 30 min
Stopping of the reaction with 100 µl/well 1 M H2SO4 and 50 mM Na2SO3
measurement of the wavelength at 490 nm and 690 nm af reference
Materials:
1 µg/ml 9E10 antibody (from different fractions)
0,5 µg/ml 9E10 antibody (from different fractions)
several stock solutions (Blocking Solution, Substrate, PBS-NKS-Phenolred)
Results:
2 active fractions of 9E10 for coulping to column NHS-activated sepharose material
Further Tasks:
Coupling of active antibodies to NHS-activated sepharose for purification of myc-tagged proteins
Preparing linearized backbones pSB1K3, pSB1A3 and pSB1T3 for ligation w/ YFP and CFP
Time: 2011-08-09,15:00-20:00
Investigators: Vanessa, Jessica, Nadine
Motivation: first step of expression backbone creation: digest linearized backbones with EcoRI and PstI
Materials:
- pSB1K3, pSB1A3 and pSB1T3
- EcoRI, PstI
- Buffer 4
- 100x BSA
- DpnI
Protocol:
Digestion protocol (following iGEM distribution protocol for linearized backbones):
- Mastermix
- 4 µl NEB Buffer 4
- 0.4 µl BSA
- 0.4 µl EcoRI
- 0.4 µl PstI
- 0.4 µl DpnI
- 14.4 µl pure water
- total: 20 µl
- reaction mix:
- 4 µl mastermix + 4 µl linearized backbone (pSB1K3, pSB1A3 or pSB1T3) from distribution
- Incubation:
- 37°C for 30 min
- Heat deactivation:
- 80°C for 20 min
- stored in fridge 4°C
Further Task:
- Digestion of YFP from pGA14mVenusGeneart and CFP from pGA14-Cerulean
- Ligation of digested linearized backbones from today with digested YFP and CFP (results: pSB1K3+YFP, pSB1T3+YFP, pSB1A2+YFP, pSB1K3+CFP, pSB1T3+CFP, pSB1A2+CFP)
- Transformation w/ ligation products, NOTE: !!! Don´t use XL1-Blue for pSB1T3+YFP and pSB1T3+CFP!!! Cells contain Tet-R already!!!
- Picking of colonies
- over-night cultures for mini-prep
- mini-prep of over-night cultures
- test-digest
- pSB1A3+YFP/CFP:
- HaeII and BglI (protocol see 2011-8-9), expected: 3 fragments
- pSB1K3+YFP/CFP
- HaeII (develop protocol, calculate exact expected bp) expected: 2 fragments
- pSB1T3+YFP/CFP
- ClaI and ApaLI (develop protocol, calculate exact expected bp) expected: 3 fragments
- also: digested, not ligated linearized backbones (pSB1K3, pSB1A3 and pSB1T3)
- also: undigested pSB1K3+YFP, pSB1T3+YFP, pSB1A2+YFP, pSB1K3+CFP, pSB1T3+CFP, pSB1A2+CFP
- if test-digest positive: digestion w/ EcoRI and XbaI
- digestion of PCR products form ??? (Ara, Lac and constitutive promotor)
- ligation of digested vectors w/digested PCR products from ???? (Ara, Lac and constitutive promotor)(results: pSB1K3+YFP+Ara, pSB1T3+YFP+Ara, pSB1A2+YFP+Ara, pSB1K3+CFP+Ara, pSB1T3+CFP+Ara, pSB1A2+CFP+Ara, pSB1K3+YFP+Lac, pSB1T3+YFP+Lac, pSB1A2+YFP+LAc, pSB1K3+CFP+Lac, pSB1T3+CFP+Lac, pSB1A2+CFP+Lac, pSB1K3+YFP+const, pSB1T3+YFP+const, pSB1A2+YFP+const, pSB1K3+CFP+const, pSB1T3+CFP+const, pSB1A2+CFP+const)
- test digestion, sequencing
Digestion of YFP from pGA14mVenusGeneart and CFP from pGA14-Cerulean
Time: 2011-08-09,15:00-20:00
Investigators: Jessica, Nadine, Vanessa
Materials:
Preparation for ligation of YFP/CFP (insert) into pSB1A3, pSB1K3 or pSB1T3 (vectors)
Materials:
- pGA14mVenusGeneart and pGA14-Cerulean
- EcoRI, PstI
- Buffer 4
- 100x BSA
Protocol:
Digestion protocol (following iGEM distribution protocol for linearized backbones):
- Mastermix
- 2 µl NEB Buffer 4
- 0.2 µl BSA
- 0.2 µl EcoRI
- 0.2 µl PstI
- 7.4 µl pure water
- total: 10 µl
- reaction mix:
- 4 µl mastermix + 4 µl 1:2 diluted DNA
- Incubation:
- 37°C for 30 min
- Heat deactivation:
- 80°C for 20 min
- YFP dig., 9.8.11, Nad & Jes stored in fridge 4°C
- CFP dig., 9.8.11, Nad & Jes stored in fridge 4°C
over night culture from PDV089
Investigators: Sandrina
Aim:
control plasmid ligation (pARW089, mdnA, geneIII --> PDV089, strategy 2)
Time: 2011-08-09,16.00-17:00
Materials/Methods:
- LB-medium with tet and amp
- cell clones from over night plate
- incubate over night at 37°C and 750 rpm
Further tasks:
- plasmid preparation and analytic digestion
Repeated PCR of mdnA for phage display (strategy 1) repeated with new ordered reversed primer and of mdnA with rfc25 restriction sites (strategy 2), gel electrophoresis and purification of PCR products
Investigators: Sandrina
Time: 2011-06-30,11:00-15:00
Aim:
- amplification of mdnA with SfiI restriction sites (vector pARW089) to clone it into pAk100 bla KDIR
- amplification of mdnA with rfc25 restriction sites to fuse it with geneIII and clone it into pARW089
Materials/Methods:
see 2011-06-10
changes:
- program: 123, Thermo Hybrid, PX2
Results:
expected bands (ca. 260 bp for strategy 1 and ca. 230 bp for strategy 2) were observed after gel electrophoresis
further tasks:
digestion of mdnA fragment with sfiI (strategy 1) and NarI and AgeI (strategy 2)
Digestion of mdnA with sfiI over night
Investigators: Sandrina
Time: 2011-08-09,16:30-17:00
Aim:
digestion of apmlificated mdnA to clone it into PAK100 bla KDIR
Materials/Methods:
50 µl sample:
- 0,2 µl BSA
- 5 µl 10x Puffer 4
- 0,5 µl sfiI
- 40 µl PCR product
- 4,3 µl H2O
incubation over night at 50°C
further tasks:
ligation with digested PAK100 bla KDIR
design primer for biobrick - redesign of pf_mdnE, pf_mdnD
Investigators: Niels, Nadine
Aim: design and order primer
Primer sequence
- pf_mdnE_EcoRI_NotI_XbaI
CAATCATCATATAACTCCGTAGATCTTCGCCGGCGCTTAAG
- pf_mdnD_EcoRI_NotI_XbaI
GTCAAAAAGGTCACGAAAGTAGATCTTCGCCGGCGCTTAAG
ordered my sigma
59th Labday 2011-08-10
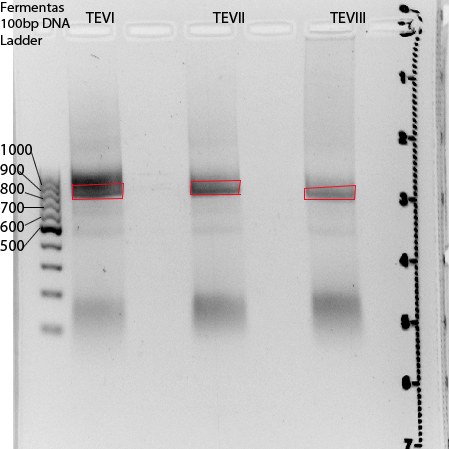
Digestion of NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragments (produced on 09.08.2011) and pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Paul
Materials:
NgoMIV_iGEM_TEV_iGEM_BamHI:
- three samples: one sample produced on 09.08.2011 and already PCR purificated; two samples produced on 09.08.2011 not PCR purificated --> PCR purification of the two unpurificated samples using "NucleoSpin ExtractII"-KIT!
TEVI:67,9ng/µl (2ml eppi in long screening reck)
TEVII:56ng/µl (2ml eppi in long screening reck)
TEVIII:68,5ng/µl (1,5 ml eppi in logn screening reck, eppi named: 2. TEV muta PCR...)
pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (contains TEV cleavage site): 470ng/µl
Digestion protocol:
1: NgoMIV_iGEM_TEV-Protease_iGEM_BamHI (3x, each sample 1x):
- 20µl NgoMIV_iGEM_TEV-Protease_iGEM_BamHI
- 1µl NgoMIV
- 1µl BamHI-HF
- 5µl 10x buffer = NEB 4
- 0.5µl 100x BSA
- 22.5µl pure water
- =50µl
2: pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (470ng/µl)
- 3µl pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (contains TEV cleavage site)
- 1µl HindIII
- 1µl BamHI-HF
- 5µl 10x buffer = NEB4
- 0.5µl 100x BSA
- 39.5µl pure water
- =50µl
-->The reaction was allowed to proceed for 2h at 37°C!
Result:
pJC354-NheI-TEV-Xho_blaFL_GGH5 vector:
Expected band: 4700bp --> Excission of band and pruification unsing "NucleoSpin ExtractII"-KIT
NgoMIV_iGEM_TEV-Protease_iGEM_BamHI:
Expected bands: 760bp --> Excission of red marked areas and pruification unsing "NucleoSpin ExtractII"-KIT
Further tasks:
Triple ligation of digested fragments and HindIII_iGEM_AraC_NgoMIV
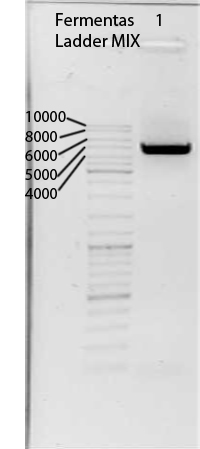
Transformation of E. coli XL1-Blue with pGA14mVenusGeneart resp. pGA14-Cerulean
Investigators: Nadine
Aim:Transformation of E. coli XL1-Blue cells with pGA14mVenusGeneart resp. pGA14-Cerulean to produce glycerol stocks for further use if necessary
Time: 2011-08-10, 9:20-?
Materials:
- pGA14mVenusGeneart resp. pGA14-Cerulean
- E. coli XL1-Blue cells
- LB medium
Method:
- addition of 1 µl plasmid to XL1-blue cells
- incubation 30 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Agarose Gel of digested pSB1K3, pSB1A3 and pSB1T3 and digested CFP and YFP fragments
Investigators: Jessica, Vanessa, Nadine
Aim:Purification of insert and vector for the first ligation of the expression backbone
Time: 2011-08-10, 9:20-16:00
Materials:
- digested pSB1K3, pSB1A3 and pSB1T3 from 2011-09-08
- digested CFP and YFP from 2011-09-08 (YFP dig. and CFP dig., 9.8.11, Nad & Jes)
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 1 % agarose gel
- 2 x 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add 1.2 µl Loading dye to each 8 µl sample
- 12 µl DNA Ladder Mix
- Running conditions: 110 V, approx. 45 min
Loading of gel 1
- gel 1: pSB1K3, pSB1A3 and pSB1T3
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker | 15 | |
| 1 | pSB1A3, EcoRI, PstI | 9.2 | 2114, 41 |
| 2 | - | - | - |
| 3 | pSB1K3, EcoRI, PstI | 9.2 | 2163, 41 |
| 4 | - | - | - |
| 5 | pSB1T3, EcoRI, PstI | - | 2422, 41 |
Result gel 1:
- bigger bands appear as expected
- 41 bp band is to small to appear in the gel
- gel extraction with Wizard SV Gel and PCR Clean-Up System (Promega):
- pSB1A3 dig. pur. 10.08.11 Nad & Jes : 9.8 ng/µl
- pSB1K3 dig. pur. 10.08.11 Nad & Jes : 10.7 ng/µl
- pSB1T3 dig. pur. 10.08.11 Nad & Jes : 11.3 ng/µl
- stored in freezer (-20°C), red box: expression backbones
Loading of gel 2
- gel 2: CFP and YFP
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | marker | ||
| 2 | from Screening (TEV Vector) | 42 | |
| 3 | - | - | - |
| 4 | CFP | 9.2 | 771, 2873 |
| 5 | - | - | - |
| 6 | YFP | 9.2 | 763, 2881 |
Result gel 2:
- in lane 4 and 6 are too many bands
- troubleshooting: it seems that the mastermixes were mixed up! In one mastermix was DpnI. DpnI digests methylated DNA. The vectors are from E. coli and therfore methylated. This could be an explanation for these band patterns.
Further tasks:
- repeat digest of YFP from pGA14mVenusGeneart and CFP from pGA14-Cerulean
Digestion of YFP from pGA14mVenusGeneart and CFP from pGA14-Cerulean
Time: 2011-08-10,15:00-17:00
Investigators: Vanessa, Jessica, Nadine
Materials:
- pGA14mVenusGeneart and pGA14-Cerulean
- EcoRI, PstI
- Buffer 4
- 100x BSA
Protocol:
Digestion protocol (following iGEM distribution protocol for linearized backbones):
- Mastermix
- 2 µl NEB Buffer 4
- 0.2 µl BSA
- 0.2 µl EcoRI
- 0.2 µl PstI
- 7.4 µl pure water
- total: 10 µl
- reaction mix:
- 4 µl mastermix + 4 µl DNA (not diluted!!!)
- Incubation:
- 37°C for 30 min
- Heat deactivation:
- 80°C for 20 min
- YFP dig., 10.8.11, Van & Jes stored in fridge 4°C
- CFP dig., 10.8.11, Van & Jes stored in fridge 4°C
Miniprep of E. coli overnight culture containing pPDV089
Investigators: Sandrina, Sabine
Aim: purification of pPDV089 for test digestion and sequencing (15 clones)
Time: 10:00-13:00
Method/Materials: see protocol 5.1 of the NucleoSpin Plasmid Kit
Further tasks: test digestion
Test digestion of ligations for strategy 2 after Plasmid preperation
Investigators: Sabine, Sandrina
Aim:control if liagation of geneIII and mdnA into pARW089 (strategy 2) worked
Time: 14:00-16:00
Materials/Methods:
Strategy 2:
- 0,5 µl XbaI
- 0,5 µl SpeI
- 2 µl 10x buffer 4 (NEB)
- 0,2 µl BSA
- 10 µl vector DNA
- 12,8 µl H2O
incubate for 1 h at 37°C
Results:
- expected size for all samples: strategy 2: ca. 5000 bp, 4000 bp, 600 bp and 200
- three different pARW089 vectors (1,2,3) were used for ligation, they were digested with the same enzymes but this was done from three different persons
- after digestion of geneIII PCR product with AatII and NgoMIV two bands were seen after gel electrophoresis, ligations were done with both samples (called here: "upper band" and "lower band")
Loading of gels
| lane | Sample | Volume in µl | Expected size in bp |
| M | marker, DNA ladder mix Fermentas | ||
| 1 | free | ||
| 2 | vector 1, upper band, clone 1 | 20 | ca. 5000, 4000, 600, 200 |
| 3 | vector 1, upper band, clone 2 | 20 | ca. 5000, 4000, 600, 200 |
| 4 | vector 1, upper band, clone 3 | 20 | ca. 5000, 4000, 600, 200 |
| 5 | vector 1, lower band, clone 1 | 20 | ca. 5000, 4000, 600, 200 |
| 6 | vector 1, lower band, clone 2 | 20 | ca. 5000, 4000, 600, 200 |
| 7 | vector 1, lower band, clone 3 | 20 | ca. 5000, 4000, 600, 200 |
| 8 | free | ||
| 9 | vector 2, upper band, clone 1 | 20 | ca. 5000, 4000, 600, 200 |
| 10 | vector 2, upper band, clone 2 | 20 | ca. 5000, 4000, 600, 200 |
| 11 | vector 2, upper band, clone 3 | 20 | ca. 5000, 4000, 600, 200 |
| 12 | vector 2, lower band, clone 1 | 20 | ca. 5000, 4000, 600, 200 |
| 13 | vector 2, lower band, clone 2 | 20 | ca. 5000, 4000, 600, 200 |
| 14 | vector 2, lower band, clone 3 | 20 | ca. 5000, 4000, 600, 200 |
| 15 | free | ||
| 16 | vector 3, upper band, clone 1 | 20 | ca. 5000, 4000, 600, 200 |
| 17 | vector 3, upper band, clone 2 | 20 | ca. 5000, 4000, 600, 200 |
| 18 | vector 3, upper band, clone 3 | 20 | ca. 5000, 4000, 600, 200 |
Further tasks:
- sequence clones 12, 13 and 17
cultivation of cells containing pAK100 bla KDIR from E. coli glycerol stocks
Investigators: Sabine, Sandrina
Aim: cultivate cells conatining pAK100 bla KDIR (vector) to purify it and use it for phage display.
Time: 2011-06-14,16:00-17.30
Materials/Methods:
- glycerol stock nr. 15, pAK100 bla KDIR, XL1-blu, 2003-07-31
- LB-medium with chloramphenicol (1:1000)
Further tasks:
plasmid preperation und digestion with SfiI
60th Labday 2011-08-11
Agarose Gel digested CFP and YFP fragments (from 2011-08-10)
Investigators: Jessica, Nadine, Vanessa
Aim:Purification of YFP and CFP fragment
Time: 2011-08-11, 8:00-11:00
Materials:
- digested CFP and YFP from 2011-09-08 (YFP dig. and CFP dig., 10.8.11, Van & Jes)
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 1 % agarose gel
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add 1.2 µl Loading dye to each 8 µl sample
- 12 µl DNA Ladder Mix
- Running conditions: 100 V, approx. 45 min
Loading of gel
- gel: CFP and YFP
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | marker | 10 | |
| 2 | - | - | - |
| 3 | CFP | 9.2 | 771, 2873 |
| 4 | - | - | - |
| 5 | YFP | 9.2 | 763, 2881 |
| 6 | - | - | - |
Result:
- bands appear as expected
- lower bands (red box) in lane 3 and 4 were excised and DNA was extracted w/ Wizard SV Gel and PCR Clean-Up System (Promega):
- CFP dig. pur. Jes & VB 11.8.2011, 5.7 ng/µl
- YFP dig. pur. Jes & VB 11.8.2011, 5.4 ng/µl
- stored in freezer (-20°C), red box: expression backbones
Further tasks:
- ligation of digested and purified YFP and CFP with digested and purified pSB1A3, pSB1K3 and pSB1T3
Ligation of digested and purified CFP and YFP fragments (from 2011-08-11) w/ digested and purified pSB1A3, pSB1K3, pSB1T3 (from 2011-08-10)
Investigators: Nadine, Vanessa, Jessica
Aim:Ligation
Time: 2011-08-11, 11:00-14:30
Materials:
- T4 Ligase
- 10x T4 Ligase buffer
- inserts
- CFP dig. pur. Jes & VB 11.8.2011, 5.7 ng/µl
- YFP dig. pur. Jes & VB 11.8.2011, 5.4 ng/µl
- vectors:
- pSB1A3 dig. pur. 10.08.11 Nad & Jes : 9.8 ng/µl,
- pSB1K3 dig. pur. 10.08.11 Nad & Jes : 10.7 ng/µl
- pSB1T3 dig. pur. 10.08.11 Nad & Jes : 11.3 ng/µl
- pure sterile water
Protocols
- to calculate the volumes http://old.gibthon.org/ was used
pSB1K3 (Volumes in µl)
| lane | CFP | YFP | Control |
| 10x T4 ligase buffer | 1 | 1 | 1 |
| T4 ligase | 1 | 1 | 1 |
| vector | 2.7 | 2.6 | 2.6 |
| insert | 5.3 | 5.4 | - |
| water | - | - | 5.4 |
pSB1A3 (Volumes in µl)
| lane | CFP | YFP | Control |
| 10x T4 ligase buffer | 1 | 1 | 1 |
| T4 ligase | 1 | 1 | 1 |
| vector | 2.8 | 2.8 | 2.8 |
| insert | 5.2 | 5.2 | - |
| water | - | - | 5.2 |
pSB1T3 (Volumes in µl)
| lane | CFP | YFP | Control |
| 10x T4 ligase buffer | 1 | 1 | 1 |
| T4 ligase | 1 | 1 | 1 |
| vector | 2.6 | 2.5 | 2.5 |
| insert | 5.4 | 5.5 | - |
| water | - | - | 5.5 |
- Incubation: 2 hrs at room temperature
Result:
- pSB1A3+YFP, lig, VB, 11.8.2011
- pSB1A3+CFP, lig, VB, 11.8.2011
- pSB1A3+control, lig, VB, 11.8.2011
- pSB1K3+YFP, lig, VB, 11.8.2011
- pSB1K3+CFP, lig, VB, 11.8.2011
- pSB1K3+control, lig, VB, 11.8.2011
- pSB1A3+YFP, lig, VB, 11.8.2011
- pSB1A3+CFP, lig, VB, 11.8.2011
- pSB1A3+control, lig, VB, 11.8.2011
- pSB1T3+YFP, lig, VB, 11.8.2011
- pSB1T3+CFP, lig, VB, 11.8.2011
- pSB1T3+control, lig, VB, 11.8.2011
- all stored in freezer (-20°V in red box: expression backbone)
Further tasks:
- Transformation in XL1-blue, except for pSB1T3+YFP/CFP: here use RV308
Ligation of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI or NgoMIV_iGEM_TEV-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV into pJC354-NheI-143C-Xho_blaFL_GGH5 or pJC354-NheI-TEV-Xho_blaFL_GGH5 vector
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Paul, Sebastian
Aim:
- 1. Triple-ligation of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (573bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-143C-Xho_blaFL_GGH5 vector (~4700bp) to create pUP_SG2_TorA_CS-14_3C_bla_AraC-14_3C
- 2.Triple-ligation of NgoMIV_iGEM_TEV_iGEM_BamHI (760bp), HindIII_iGEM_AraC_NgoMIV (1273bp) and pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (~4700bp)to create pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
Calculation of volumes to be used with: [http://www.gibthon.org/ligate.html ligation calculator] with 1:1 molar ratio
Materials:
14_3C: 2 reaction batches (we have two digested pJC354-NheI-143C-Xho_blaFL_GGH5 vector fractions)
1:
- 2 µL NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI fragment (573bp, 3.2ng/µl)
- 1,5 µL AraC fragment
- 4,5 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector (8.3ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
2:
- 1,2 µL NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (573bp, 3.2ng/µl) fragment
- 0,8 µL AraC fragment
- 6 µL pJC354-NheI-143C-Xho_blaFL_GGH5 vector (6.7ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
- 2 controls: As 1 and 2 but with water instead of fragment
TEV: 3 reaction batches (we have three digested NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment fractions)
1:
- 0.8 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 18.5ng/µl) (TEVI, see 10.08.2011)
- 2,6 µL AraC fragment
- 4,6 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (19.4ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
2:
- 1.1 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 12.5ng/µl) (TEVII, see 10.08.2011)
- 2,5 µL AraC fragment
- 4,4 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (19.4ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
3:
- 0.7 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 20.4ng/µl) (TEVIII, see 10.08.2011)
- 2,6 µL AraC fragment
- 4,7 µL pJC354-NheI-TEV-Xho_blaFL_GGH5 vector (19.4ng/µl)
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
- 1 control: As 2 but with water instead of fragment
Used method:
ligation at room temperatur for 1h
Further going:Transformation of XL1blue cells with ligation products
Transformation of E. Coli XL1 Blue Cells with ligation products (NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI or NgoMIV_iGEM_TEV-Protease_iGEM_BamHI, HindIII_iGEM_AraC_NgoMIV into pJC354-NheI-143C-Xho_blaFL_GGH5 or pJC354-NheI-TEV-Xho_blaFL_GGH5 vectors)
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators:Paul, Sascha
Aim:transform the ligation into E. coli XL1 blue
Materials:
8x XL1 blue cells from -80 stock, 2x 14_3C-ligations + 2x controls ; 3x TEV-ligations + 1x control
protocol:
- Taw cells on ice
- addition of 10 µl ligation reaction to cells (XL1-blue, tet-resistance, 60µl) in 2 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C for 60 min in Eppendorf thermomixer at 600 rpm
- plating on LB medium with 1,5 % agar, 100 µg/ml chloramphenicol,
- storage over night at 37°C
Results
No colonies in case of 14_3C Protease, no colonies in controls of 14_3C
Tev: three colonies on each plate, including control plate
Further tasks
Picking colonies, making precultures and colony-PCRs and isolation/sequencing of pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
Transformation
Time: 2011-8-11, 14:30 - 17:30
Investigators: Vanessa, Jessica, Nadine, Niels
Material:
- pSB1A3+YFP, lig, VB, 11.8.2011
- pSB1A3+CFP, lig, VB, 11.8.2011
- pSB1A3+control, lig, VB, 11.8.2011
- pSB1K3+YFP, lig, VB, 11.8.2011
- pSB1K3+CFP, lig, VB, 11.8.2011
- pSB1K3+control, lig, VB, 11.8.2011
- pSB1A3+YFP, lig, VB, 11.8.2011
- pSB1A3+CFP, lig, VB, 11.8.2011
- pSB1A3+control, lig, VB, 11.8.2011
- pSB1T3+YFP, lig, VB, 11.8.2011
- pSB1T3+CFP, lig, VB, 11.8.2011
- pSB1T3+control, lig, VB, 11.8.2011
- 6 x XL1-blue cells (competent) for pSB1A3 and pSB1K3 variants
- 3 x RV380 cells (competent) for pSB1T3 variants
Method:
- addition of 2 µl ligation reaction to XL1-blue cells or RV380 cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin
- plating on agar plates containing 100 µg/ml kanamycin
- plating on agar plates containing 100 µg/µl ampicillin
- incubation over night at 37°C
NOTE:
- pSB1T3+YFP/CFP have the same label; we have to check the insert with test digestion
Further tasks:
control cell clones tomorrow morning
Repeated PCR gene III for phage display (strategy 2)
Investigator: Sabine
Time: 2011-08-11,1:00-12:30
Aim:
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_geneIII_NgoMIV_XbaI_myc and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl pak100blaKDIR
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
Purification:
- NucleoSpin Extract II
Further tasks:
- digestion
Miniprep of E. coli overnight culture containing pakblaKDIR
Investigators: Sabine
Aim: purification of pakblaKDIR
Time: 10:00-11:00
Method/Materials: see protocol 5.1 of the NucleoSpin Plasmid Kit
Further tasks:
- digestion with Sfi (strategy I)
- PCR of geneIII (strategy II)
digestion of pak blaKDIR (strategy 1)
Investigator: Sandrina, Sabine
Aim: ligation of digested mdnA into pak blaKDIR to get phage display vector pPDV100 (strategy I)
Time: 2011-08-11, 11:00-14:00
reaction components:
- 15 µl pak blaKDIR (ca 1 µg)
- 2 µl NEB 10x buffer
- 0,2 µl 100x BSA
- 1 µl restriction enzyme SfiI
- 1,8 µl water
reaction conditions:
- 3 h, 37°C
sending clone 12, 13 and 17 from ligation strategy 2 (see 2011-08-10) to MWG for sequencing
Investigators: Sandrina, Sabine
Time: 2011-8-11, 12:00 - 15:00
Aim: get sequence of generated phage display vector pPDV089 (strategy 2) to control the ligation of digested pARW089 with digested mdnA and gene III
Method/Materials:
- 50-100 ng DNA in 15 µl sample
- used primer: sf_mdna_1 (nr. 6)
Further tasks:
perform alignment
Gel electrophoresis of digested PAK100 bla KDIR vector(strategy 1) and PCR of geneIII
Investigators: Sandrina
Aim:Purification of PAK100 bla KDIR (strategy 1) and analysis if PCR worked
Time: 2011-08-11,15:00-16:30
Materials/Methods:
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- digested vector PAK100 bla KDIR
- DNA Ladder Mix(Fermentas)
- 6x Loading Dye (Fermentas)
Method:
1. Production of a 1 % agarose gel
- adding 2 µl gel red
2. Run
- 100 V
- time: 00:45 h
Results:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | marker | ||
| 1 | geneIII PCR | 5 | ca. 400 |
| 2 | digested PAK100 bla KDIR vector | 30 | ca. 5500, 800 |
one band was cutted out of the gel for purifacation
Further tasks:
ligation of mdnA with pAK100 bla KDIR
ligation of mdnA into pak100blaKDIR (stategy 1)
Investigator: Niels, Sandrina
Aim: generate phage display vector pPDV100 (strategy 1)
Time: 2011-07-20,16:30-18:00
Method/Materials:
- 3 µl (ca 90 ng) Sfi-digested vector pak100
- 1 µl (ca 1000 ng) Sfi-digested PCR fragment mdnA from over night digested sample (2011-08-09)
- 2 µl 10x T4 Ligase Buffer
- 1 µl T4 Ligase
- 13 µl water
- incubate over night at 14°C
Further Tasks: transformation of competent cells
61th Labday 2011-08-12
Ligation of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI or NgoMIV_iGEM_TEV-Protease_iGEM_BamHI with HindIII_iGEM_AraC_NgoMIV
Investigators: Sebastian, Paul
Aim:
1. Ligation of NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI (573bp) and HindIII_iGEM_AraC_NgoMIV (1273bp)
2. Ligation of NgoMIV_iGEM_TEV_iGEM_BamHI (760bp) and HindIII_iGEM_AraC_NgoMIV (1273bp)
3. Amplification of ligated fragments via PCR
Calculation of volumes to be used with: [http://www.gibthon.org/ligate.html ligation calculator] with 1:1 molar ratio
Materials:
1:
- 4.3 µL NgoMIV_iGEM_14_3C-Protease_iGEM_BamHI fragment (573bp, 3.2ng/µl)
- 3.7 µL AraC fragment
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
2:
- 1.1 µL NgoMIV_iGEM_TEV-Protease_iGEM_BamHI fragment (760bp, 18.5ng/µl) (TEVI, see 10.08.2011)
- 4 µL AraC fragment
- 2.9 µL pure water
- 1 µL T4 liagtion buffer (Fermentas)
- 1 µL T4 ligase (Fermentas)
- =10µl
Used method:
ligation at room temperatur for 1h
3:
- 10µl of ligation reaction batch
- 1µl dNTP
- 2.5µl of primer1
- 2.5µl of primer2
- 5µl 25mM MgCl2
- 5µl Amplification Buffer 10x (genaxxon)
- 0.5µl Taq-Polymerase (genaxxon)
Primer:
TEV+AraC:
r_Tev_iGEM_BamHI
f_AraC_HindIII_iGEM
14_3C+AraC:
r_14_3C_iGEM_BamHI
f_AraC_HindIII_iGEM
Used method:
- Initial Denat: 3min 94°C
- 25x
- Denat: 2.02min 94°C
- Anneal: 2.02min 70°C
- Extension: 2.02min 72°C
- final extension: 10min 72°C
Results:
No results, no expected bands were visible
Further Tasks:
resolving of PCR products on preparative 1% agarose gel and exciccion of correspoinding bands (1:~1800bp, 2:~2000bp)
File:UP AG 1% 2011-08-12 prep-TEV AraC.jpg
Colony PCR of TEV clones obtained from transformation
Stefan, Sebastian
Aim:
get a positive clone
Materials:
- Primer:
- f_AraC_HindIII_iGEM
- r_TEV_iGEM_BamHI
- 2.5 µL forward Primer (25 pmol)
- 2.5 µL reverse Primer (25 pmol)
- 5 µL 10x Polymerase buffer (Genaxxiom)
- 5 µL MgCL2
- 1 µL dNTPs
- 0.5 µL Taq Polymerase (Genaxxiom)
- 33.5 µL H20
10 colonies were picked
Used method:
- Initial Denat: 3 min 94°C
- 25x
- Denat: 2.02 min 94°C
- Anneal: 2.02 min 70°C
- Extension: 2.02 min 72°C
- final extension: 10min 72°C
Further Tasks:
gel electrophoresis of PCR products 1% agarose gel of corresponding bands (1:~1800bp, 2:~2000bp)
Results of Transformation from 2011-8-11 and Overnight Cultures of marked colonies
Time: 2011-8-12, 9:00-9:30
Investigators : Jessica, Nadine, Katharina
Aim: check Transformation
Results:
Preparing Overnight Cultures of marked colonies
Time: 2011-8-12, 18:00-18:30
Investigators : Jessica
Materials/Method:
- 5 ml LB-Medium with 5 µl antibiotic (either Tet, Amp or Kan)
- cultures:
- pSB1T3+YFP I clone I 12.08.11 Jes, pSB1T3+YFP I clone II 12.08.11 Jes, pSB1T3+YFP I clone III 12.08.11 Jes
- pSB1K3+YFP clone I 12.08.11 Jes, pSB1T3+YFP clone II 12.08.11 Jes, pSB1T3+YFP clone III 12.08.11 Jes
- pSB1K3+CFP clone I 12.08.11 Jes, pSB1T3+CFP clone II 12.08.11 Jes, pSB1T3+CFP clone III 12.08.11 Jes
- pSB1A3+CFP clone I 12.08.11 Jes, pSB1A3+CFP clone II 12.08.11 Jes
- pSB1A3+CFP clone I 12.08.11 Jes, pSB1A3+CFP clone II 12.08.11 Jes
Further Task:
- repeat Transformation of pSB1T3+YFPII and pSB1A3+YFP
- do miniprep of overnight cultures and confirm insert by digest (see 2011-08-09)
Transformation of generated pPDV100 from over night ligation (strategy 1)
Investigators: Sabine, Sandrina
Aim:amplification of pPDV100
Time: 10:00-12:00
Method:
- addition of 10 µl ligation reaction to XL1-blue cells
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml tetracyclin and 100 µg/µl ampicillin
- storage over night at 37°C
Further tasks:
control cell clones through test digestion with sfiI
Repeated PCR gene III for phage display (strategy 2)
Investigator: Sandrina, Sabine
Time: 2011-08-12,12:30-17:00
Aim:
- amplificate GeneIII with NgoMIV and AatII restriction sites (strategy 2)
Primer:
- primer: pf_geneIII_NgoMIV_XbaI_myc and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 5 µl pak100blaKDIR
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 37,75 µl DNase free water
Purification:
- NucleoSpin Extract II
Further tasks:
- digestion with AatII and NgoMIV
Miniprep of overnight cultures of Cerulean and mVenus Geneart
Investigators: Katharina
Time: 2011-08-12, 9:00-10:00
Aim:
1. Miniprep:
- 2 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 30 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| Cerulean | 530.1 |
| mVenus Geneart | 573.6 |
- stored in -20°C (red box, expression backbones)
2. Preparation of glycerol stocks:
- adding 300 µl glycerol to 700 µl culture
gel electrophoresis of colony PCR(TEV)
Investigators: Sascha, Paul, Sebastian
Aim:
screen for positiv clone
Result:
Gel electrophoresis of PCR products (5µl) on 1.5% and agarose gels, respectively.
Expected band (AraC + TEV): ~ 2000 bp.
One positive clone TEV 3 III.
Set up a pre-culter of clon TEV 3 III.
Further tasks:
Stock culture, plasmid preparation
Transformation of pSB1A3+YFP in XL1-Blue and pSB1T3+YFPII in RV308
Investigators: Katharina
Aim: Transformation of Ligations
Time: 2011-08-12, 10:00-13:00
Materials:
- competent E. coli cells (XL1-Blue and RV308, respectively)
- ligation products: pSB1A3+YFP, lig, VB, 11.8.2011 and pSB1T3+YFP II, lig, VB, 11.8.2011
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue, RV) in 1.5 ml Eppi,
- incubation 25 min on ice,
- heat shock 45 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 80 min,
- plating on LB medium with appropriate antibiotic (Amp and Tet,respectively
- storage over night at 37°C
- 2 plates: pSB1T3+YFP II Jes and pSB1A3+YFP Jes
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Design and ordering of primers to produce BioBricks of the mdn genes
Investigators: Jessica, Nadine, Katharina
Time: 2011-08-12, 11:00-14:00
Materials:
- Geneious
Results:
- pf_mdnABC_EcoRI_NotI_XbaI 12.08. : TTAATGAATTCGCGGCCGCTTCTAGATGGCATATCCCAACGATC
- pf_mdnB_EcoRI_NotI_XbaI 12.08.: ATTATGAATTCGCGGCCGCTTCTAGATGAAAGAATCGCCTAAAGTTG
- pf_mdnC_EcoRI_NotI_XbaI 12.08.: TATTTGAATTCGCGGCCGCTTCTAGATGACCGTTTTAATTGTTAC
- pf_mdnD_EcoRI_NotI_XbaI 12.08.: TATATGAATTCGCGGCCGCTTCTAGATGAAAGCACTGGAAAAACTG
- pf_mdnE_EcoRI_NotI_XbaI 12.08.: TAAATGAATTCGCGGCCGCTTCTAGATGCCTCAATATACTACTAAAC
- pr_mdnABC_SpeI_NotI_PstI 12.08.: ATTTCTGCAGCGGCCGCTACTAGTATTATGAGTTAACTAGGATTTC
- pr_mdnB_SpeI_NotI_PstI 12.08.: TAATCTGCAGCGGCCGCTACTAGTAACTAAAAAATCAGCGATCGC
- pr_mdnD_SpeI_NotI_PstI 12.08.: ATTTCTGCAGCGGCCGCTACTAGTATCAGCAAACCCTACTTAATTTC
- pr_mdnE_SpeI_NotI_PstI 12.08.: ATTTCTGCAGCGGCCGCTACTAGTACTATATTCTCACCCATTTTAAG
Further tasks:
- develop PCR program
- PCR
62th Labday 2011-08-13
Mini-Prep of pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV and creation of glycerol stock culture
Investigator: Sebastian
Aim:
- Isolation of created plasmid pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV and establishing of an E. coli Xl1 blue glycerol stock culture for later use
Materials:
- Overnight culture of E. coli XL1 Blue transformed with pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV plasmid
- NucleoSpin Plasmid (NoLid) Kit purchased by Macherey-Nagel
Method:
preparing the stock culture
- 350 µl sterile glycerol where mixed with 350 µl form the overnight culture of E. coli XL1 blue transformed with plasmid
pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV, vortexed and stored at -80°C (stock number: G1)
Mini-prep of pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
- preparation was performed as described in the manual of the used Kit
Results:
- glycerol stock culture G1: E. coli (XL1 blue) transformed with pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
- isolated plasmid pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
Further Tasks:
- sequencing of the plasmid pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
- growth test for the stock culture at different conditions (with induction by IPTG and arabinose at increasing ampicillin concentrations) --> "survival test"
- preparing of competent cells, which are transformed with pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
Miniprep of pSB1X3 + Y (X - A/T/K ; Y - YFP/CFP)
Investigators: Niels
Aim: isolate plasmid of pSB1X3 + Y (X - A/T/K ; Y - YFP/CFP)
Material: 5 ml / over night culture
- pSB1T3+YFP I clone I 12.08.11 Jes
- pSB1T3+YFP I clone II 12.08.11 Jes
- pSB1T3+YFP I clone III 12.08.11 Jes
- pSB1K3+YFP clone I 12.08.11 Jes
- pSB1T3+YFP clone II 12.08.11 Jes
- pSB1T3+YFP clone III 12.08.11 Jes
- pSB1K3+CFP clone I 12.08.11 Jes
- pSB1T3+CFP clone II 12.08.11 Jes
- pSB1T3+CFP clone III 12.08.11 Jes
- pSB1A3+CFP clone I 12.08.11 Jes
- pSB1A3+CFP clone II 12.08.11 Jes
<b>Method:
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl elution buffer2O
Check concentration with NanoDrop
- pSB1T3+YFP I clone I 12.08.11 Jes 10,3 ng/µl
- pSB1T3+YFP I clone II 12.08.11 Jes 16,6 ng/µl
- pSB1T3+YFP I clone III 12.08.11 Jes 24,2 ng/µl
- pSB1K3+YFP clone I 12.08.11 Jes 29,7 ng/µl
- pSB1K3+YFP clone II 12.08.11 Jes 22,9 ng/µl
- pSB1K3+YFP clone III 12.08.11 Jes 25,7 ng/µl
- pSB1K3+CFP clone I 12.08.11 Jes 23,4 ng/µl
- pSB1K3+CFP clone II 12.08.11 Jes 37,6 ng/µl
- pSB1K3+CFP clone III 12.08.11 Jes 27,8 ng/µl
- pSB1A3+CFP clone I 12.08.11 Jes 5,8 ng/µl
- pSB1A3+CFP clone II 12.08.11 Jes 21,7 ng/µl
<b>NOTE:
- pSB1T3+YFP/CFP have the same label; we have to check the insert with test digestion
Further tasks:
- digest
63th Labday 2011-08-14
Repeated PCR of mdnA and gene III for phage display (strategy 1+2)
Investigator: Sandrina, Sabine
Time: 2011-08-14,11:00-14:00
Aim:
- amplification of mdnA with SfiI restriction sites (strategy 1)
- amplification of mdnA with NarI and AgeI restriction sites (strategy 2)
- amplification of GeneIII with NgoMIV and AatII restriction sites (strategy 2)
- 5 PCRs of every gene for testing different digestion and ligation conditions
Primer:
- primer 31 and 56: pf_sfi-mdnA_2 and pr_sfi_mdnA_myc-2 (mdnA, strategy 1)
- primer 19 and 32: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII (mdnaA, strategy 2)
- primer 54 and 55: pf_geneIII_NgoMIV_XbaI_myc and pr_geneIII_iGEM_AatII (geneIII, strategy 2)
Reaction Components:
- 2 µl (ca 16 ng) Vector pARW089 (mdnA) or pakblaKDIR (geneIII)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 39,75 µl water
PCR condition changes for geneIII:
- higher annealing temperature: 60°C
- Stage 1: only 10 cycles (instead of 15)
- purification of PCR fragments with QIAquick Gel Extraction Kit (250)
Further tasks:
- digestion
Results:
PCRs of mdnA and mdnA were ok, PCR of geneIII: only oligo band
digestion of pak blaKDIR (strategy 1)
Investigator: Sandrina, Sabine
Aim: cut geneIII out of pak100 blaKDIR to use it as template for PCR of geneIII
Time: 2011-08-14, 12:00-15:00
reaction components:
- 5 µl pak blaKDIR (ca 350 ng)
- 2 µl NEB 10x buffer 2
- 1 µl restriction enzyme AvaI
- 1 µl restriction enzyme HindIII
- 11 µl water
reaction conditions:
- 3 h, 37°C
overnight culture of 10 picked clones of XL blue cells transformed with pPDV100
Investigators: Sandrina, Sabine
Aim: amplification and purification of generated phage display vector pPDV100 for test digestion and sequencing
Time: 16:00-16:30
Method/Materials:
5 ml LB medium per clone containining 100 µg/ml chloramphenicol
storage over night at 37°C and 800 rpm
Further tasks:
plasmid preparation, test digestion and sequencing
generate over night culture of
Investigators: Niels
Aim: generate a over night culture for isolating the plasmid
Material:
- 5 ml LB Medium
- 5 µl ampicilin (20 mg/ml)
- plate pSB1A3+YFP
- clone I 12.08.11 Jes
- clone II 12.08.11 Jes
- clone III 12.08.11 Jes
further tasks
- isolation of plasmid
- digest with
64th Labday 2011-08-15
Miniprep of pSB1A3 + YFP clone I, pSB1A3 + YFP clone II, pSB1A3 + YFP clone III
Investigators: Steffi
Time: 2011-08-15, 07:00-10:00
Aim: isolate plasmid of pSB1A3 + YFP clone I, pSB1A3 + YFP clone II, pSB1A3 + YFP clone III
Material: 5 ml over night culture
- pSB1A3+YFP I clone I 12.08.11 Jes
- pSB1A3+YFP I clone II 12.08.11 Jes
- pSB1A3+YFP I clone III 12.08.11 Jes
Method:
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl elution buffer2O
Check concentration with NanoDrop and Agarosegel
- pSB1A3+YFP I clone I: 134.3 ng/µl
- pSB1A3+YFP I clone II: 113.8 ng/µl
- pSB1A3+YFP I clone III: 179.2 ng/µl
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 6 | |
| 1 | - | - | |
| 2 | pSB1A3+YFP I clone I 12.08.11 Jes | 1 | |
| 3 | pSB1A3+YFP I clone II 12.08.11 Jes | 1 | |
| 4 | pSB1A3+YFP I clone III 12.08.11 Jes | 1 |
Further tasks:
- digest
Test digest of minipreps of pSB1X3+YFP/CPF from 2011-08-13 and 2011-08-15
Investigators: Jessica, Laura, Steffi, Vanessa
Time: 2011-08-15, 10:00-15:00
Aim: prove of Insert YFP/CPF
Materials:
pSB1T3+YFP I clone I 12.08.11 Jes pSB1T3+YFP I clone II 12.08.11 Jes pSB1T3+YFP I clone III 12.08.11 Jes pSB1K3+YFP clone I 12.08.11 Jes pSB1T3+YFP clone II 12.08.11 Jes pSB1T3+YFP clone III 12.08.11 Jes pSB1K3+CFP clone I 12.08.11 Jes pSB1T3+CFP clone II 12.08.11 Jes pSB1T3+CFP clone III 12.08.11 Jes pSB1A3+CFP clone I 12.08.11 Jes pSB1A3+CFP clone II 12.08.11 Jes
- ApaLI, ClaI, HaeII, BglI
- NEB Buffer 4
- NEB Buffer 2
- 100x BSA
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
- 6x Loading Dye (Promega)
Digestion protocol for pSB1K3+YFP/CFP (with HaeII)
- 2 µl DNA
- 0.5 µl HaeII
- 2 µl 10x buffer = NEB 4
- 15.8 µl pure water
- total: 20µl
Digestion protocol for pSB1A3+YFP/CFP (with HaeII, BglI)
- 2 µl DNA
- 0.5 µl HaeII
- 0.5 µl BglI
- 2 µl 10x buffer = NEB 2
- 0.2 µl 100xBSA
- 15.3 µl pure water
Digestion protocol for pSB1T3+YFPI (with ApaLI, ClaI)
- 2 µl DNA
- 0.5 µl ApaLI
- 0.5 µl ClaI
- 2 µl 10x buffer = NEB 4
- 0.2 µl 100xBSA
- 15.3 µl pure water
37°C for ~1h
Production of one 1 % agarose gel
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to each gel
Loading gels and running
- Add 4 µl Loading dye to each 20 µl sample
- 12 µl DNA Ladder Mix
- Running conditions: 100 V, approx. 45 min
Loading of gels
Gel 1
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 12 | |
| 1 | pSB1A3+YFP clone I | 24 | 765, 924, 1188 |
| 2 | - | ||
| 3 | pSB1A3+YFP clone II | 24 | 765, 924, 1188 |
| 4 | pSB1A3+YFP clone III | 24 | 765, 924, 1188 |
| 5 | pSB1A3+CFP clone I | 24 | 765, 924, 1188 |
| 6 | pSB1A3+CFP clone II | 24 | 765, 924, 1188 |
Gel 2
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 12 | |
| 1 | pSB1K3+YFP clone I | 24 | 924, 2010 |
| 2 | pSB1K3+YFP clone II | 24 | 924, 2010 |
| 3 | pSB1K3+CFP clone I | 24 | 924, 2010 |
| 4 | pSB1K3+CFP clone II | 24 | 924, 2010 |
| 5 | pSB1K3+CFP clone III | 24 | 924, 2010 |
| 6 | pSB1K3+YFP clone III | 24 | 924, 2010 |
| 7 | - | ||
| 8 | pSB1T3+YFP I clone I | 24 | 536, 869, 1788 |
| 9 | pSB1T3+YFP I clone II | 24 | 536, 869, 1788 |
| 10 | pSB1T3+YFP I clone III | 24 | 536, 869, 1788 |
Result:
- Inserts couldn't be confirmed
- possible explanation: concentration of insert to low before ligation
Further tasks:
- repeat digest of pGA14mVenusGeneart and pGA14-Cerulean to get YFP and CFP
- repeat ligation
Transformation of E. coli RV308 with pSB1T3+YFPII
Investigators: Niels, Jessica, Steffi
Aim:Transformation of E. coli RV - cells with
Materials:
- ligation of pSB1T3+YFPII from 2011-08-11
- E. coli RV308
- LB medium
Method:
- addition of 2 µl plasmid to RV - cells
- incubation 30 min on ice,
- heat shock 90 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 75 min at 37 °C and 700 rpm
- plating on agar plates containing 100 µg/µl tetracycline
- storage over night at 37°C
Further tasks:
- Picking clones for overnight culture
mini prep and test digestion of phage display vector pPDV100 from over night cultures (strategy 1)
Investigators: Laura, Sabine
Aim: test digestion of pPDV100 to control mdnA cloning into pak blaKDIR
Time: 2011-08-15,12:00-14:15 and 16.00-18.00
Digestion of vector pPDV100 with Sfi (10 clones)
- 10 µl sample
- 2 µl NEB 10x buffer 4
- 1 µ restriction enzyme SfiI
- 7 µl water
- over night at 50°C
Digestion of vector pPDV100 with PvuII (10 clones)
- 10 µl sample
- 2 µl NEB 10x buffer 4
- 1 µ restriction enzyme PvuII
- 7 µl water
- over night at 37°C
digestion of vector pARW089 (strategy 2)
Investigators: Laura, Sabine
Aim: cloning od mdnA/geneIII fusion gene into pARW089
Time: 2011-08-15,17:30-18:00
Reaction components:
- 5 µl pARW089
- 2 µl NEB 10x buffer 4
- 1 µ restriction enzyme NarI
- 1 µ restriction enzyme AatII
- 11 µl water
Reaction conditions:
- over night at 37°C
Results:
- no of the expected bands (10,2 kb and 160 bp)
Survival test for E. coli XL1 blue transformed with pUP_SG1_ssTorA_CS-TEV_bla_AraC-TEV
Investigator: Sebastian, Stefan, Sascha
Aim:
- testing the influence of the induction with IPTG and arabinose (different concentrations)
- capacity of resistence vs. ampicillin after induction of TorA_CS-TEV_bla with IPTG
Materials:
- LB-Agar
- deluted overnight culture of E. coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
- LB-Media
- Stock solutions of 1M IPTG, 1M arabinose (ara), 100mg/ml ampicillin (amp) and 25 mg/ml chloramphenicol (cm)
Methode:
100µl of deluted overnight culture of E. coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV (OD (600 nm)=0.002) were plated on different perpared agar plates and incubated over night at 30°C.
- used plates:
- total plates: 15
- total plates: 15
- 1 plate with cm (25µg/ml)
- 1 plate with cm (25µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG
- 1 plate with cm (25µg/ml), 1 mM IPTG
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (50 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (50 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (100 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (100 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (200 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (200 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (400 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (400 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (800 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (800 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM ara
- 1 plate with cm (25µg/ml), 1 mM ara
- 1 plate with cm (25µg/ml), 5 mM ara
- 1 plate with cm (25µg/ml), 5 mM ara
- 1 plate with cm (25µg/ml), 10 mM ara
- 1 plate with cm (25µg/ml), 10 mM ara
- 1 plate with cm (25µg/ml), 20 mM ara
- 1 plate with cm (25µg/ml), 20 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 1 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 1 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 5 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 5 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 10 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 10 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 20 mM ara
- 1 plate with cm (25µg/ml), 1 mM IPTG, 20 mM ara
Survival test for E.coli XL1 blue transformed with pUP_SG2_ssTorA_CS-Pre_bla
Investigator: Sebastian, Stefan, Sascha
Aim:
- testing the influence of the induction with IPTG and arabinose (different concentrations)
- capacity of resistence vs. ampicillin after induction of TorA_CS-TEV_bla with IPTG
Materials:
- LB-Agar
- deluted overnight culture of E.coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV
- LB-Media
- Stock solutions of 1M IPTG, 1M arabinose (ara), 100mg/ml ampicillin (amp) and 25 mg/ml chloramphenicol (cm)
Methode:
100µl of deluted overnight culture of E.coli XL1-blue transformed with pUP_SG1_TorA_CS-TEV_bla_AraC-TEV (OD (600 nm)=0.002) were plated on different perpared agar plates and incubated over night at 30°C.
- used plates:
- total plates: 15
- total plates: 15
- 1 plate with cm (25µg/ml)
- 1 plate with cm (25µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG
- 1 plate with cm (25µg/ml), 1 mM IPTG
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (50 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (50 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (100 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (100 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (200 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (200 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (400 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (400 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (800 µg/ml)
- 1 plate with cm (25µg/ml), 1 mM IPTG, amp (800 µg/ml)
Results:
Further tasks:
Results:
Further tasks:
65th Labday 2011-08-16
Digestion of YFP from pGA14mVenusGeneart and CFP from pGA14-Cerulean
Time: 2011-08-16,08:00-14:00
Investigators: Steffi
Materials:
Preparation for ligation of YFP/CFP (insert) into pSB1A3, pSB1K3 or pSB1T3 (vectors)
Materials:
- pGA14mVenusGeneart and pGA14-Cerulean
- EcoRI-HF, PstI-HF
- Buffer 4
- 100x BSA
Protocol:
Digestion protocol (following iGEM distribution protocol for linearized backbones):
- Mastermix
- 2 µl NEB Buffer 4
- 0.2 µl BSA
- 1 µl EcoRI-HF
- 1 µl PstI-HF
- 10,8 µl pure water
- total: 15 µl
- reaction mix:
- 15 µl mastermix + 5 µl DNA
- Incubation:
- 37°C for 1 h
- Heat deactivation:
- 80°C for 20 min
Further Task:
- gel electrophoresis
- gel extraction and purification
Agarose Gel digested CFP and YFP fragments
Investigators: Steffi, Jessica, Nicole
Aim:Purification of YFP and CFP fragment
Time: 2011-08-16, 11:00-12:00
Materials:
- digested CFP and YFP
- Agarose broad range (Roth)
- 1x TAE buffer
- Gel Red
- DNA Ladder Mix (Fermentas)
- 6x Loading Dye (Fermentas)
Production of one 1 % agarose gel
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red gel
Loading gel and running
- Add 5 µl Loading dye to each 20 µl sample
- 6 µl DNA Ladder Mix
- Running conditions: 85 V, approx. 1 h
Loading of gel
- gel: CFP and YFP
| lane | Sample | Volume in µl | Expected size in bp |
| 1 | marker | 6 | |
| 2 | - | - | - |
| 3 | CFP | 20 | 771, 2873 |
| 4 | - | - | - |
| 5 | YFP | 20 | 763, 2881 |
| 6 | - | - | - |
Result:
- bands appear as expected
- lower bands (red box) in lane 3 and 4 were excised and DNA was extracted w/ Wizard SV Gel and PCR Clean-Up System (Promega):
- CFP dig. pur. Jes & VB 11.8.2011, ???? ng/µl
- YFP dig. pur. Jes & VB 11.8.2011, ???? ng/µl
Further tasks:
- ligation of digested and purified YFP and CFP with digested and purified pSB1A3, pSB1K3 and pSB1T3
Sequencing of pSB1A3 and pSB1K3 carrying Cerulean resp. mVenus
Investigators:Nicole
Time: 2011-08-16, 13:00-14:00
Aim: Confirmation of ligation of linearized backbones pSB1A3 and pSB1K3 with mVenus and Cerulean as dummies
Materials:
- pSB1A3 carrying Cerulean, miniprep done by Nadja/Nicole, cDNA = 164,1 ng/ µl
- pSb1A3 carrying mVenus, miniprep done by Steffi, cDNA = 179,2 ng/ µl
- pSB1K3 carrying Cerulean, miniprep done by Nadja/Nicole, cDNA = 216,2 ng/ µl
- pSb1K3 carrying mVenus, miniprep done by Nadja/Nicole, cDNA = 178,9 ng/ µl
- Sequencing primer: VF2
- Freelabels for Value Read Tube (MWG Eurofins)
Method:
File:UP 2011-08-16 pSB1A3 pSB1K3 CFP YFP VF2.jpg
- DNA concentration (for sequencing): 70 ng/ µl
- Total volume: 15 µl
- Primer concentration: 2 pmol/ µl
- Total volume: 50 µl
- sent to MWG Eurofins
Results:
- expected 2011-08-18, afternoon
Further tasks:
- Analyzing sequencing results
Planning and accomplishing the control PCRs of pSB1A3 and pSB1K3 carrying mVenus resp. Cerulean
Investigators: Jessica, Nicole
Time: 2011-08-16, 14:00-16:00
Aim: Confirmation of ligation of linearized backbones pSB1A3 and pSB1K3 with mVenus and Cerulean as dummies; therefore perfoming PCRs using VF2 and VR2 primers, which bind in the backbone and lead to amplification of insert
Materials:
1. Plasmids
- pSB1A3 carrying Cerulean, miniprep done by Nadja/Nicole, cDNA = 164,1 ng/ µl
- pSb1A3 carrying mVenus, miniprep done by Steffi, cDNA = 179,2 ng/ µl
- pSB1K3 carrying Cerulean, miniprep done by Nadja/Nicole, cDNA = 216,2 ng/ µl
- pSb1K3 carrying mVenus, miniprep done by Nadja/Nicole, cDNA = 178,9 ng/ µl
2. Primer
- VF2
- VR2
- bind in the backbone and lead to amplification of insert
3. Other materials
- Polymerase S and corresponding buffer
Method:
1. Reaction mix (volumes in µl)
| Ingredient | pSB1A3+CFP | pSB1A3+YFP | pSB1K3+CFP | pSB1K3+YFP |
|---|---|---|---|---|
| DNA (1:100) | 6.0 | 5.6 | 4.6 | 5.6 |
| Buffer (15 mM MgCl2) | 5.0 | 5.0 | 5.0 | 5.0 |
| dNTPs (10 mM each) | 1.0 | 1.0 | 1.0 | 1.0 |
| MgCl2 (25 mM) | 2.0 | 2.0 | 2.0 | 2.0 |
| VF2 (10 mM) | 1.0 | 1.0 | 1.0 | 1.0 |
| VR2 (10 mM) | 1.0 | 1.0 | 1.0 | 1.0 |
| Genaxxon Polymerase S | 0.3 | 0.3 | 0.3 | 0.3 |
| H2O | 33.7 | 34.1 | 35.1 | 34.1 |
2. PCR program (IGCONT1)
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 94°C | Hold |
| Initial denaturation | 94°C | 3 min |
| Denaturation | 94°C | 45 s |
| Annealing | 50°C | 45 s |
| Extension | 72°C | 70 s |
| Final extension | 72°C | 10 min |
- 30 cycles of denaturation, annealing and extension
Agarose gel electrophoresis
| lane | Sample | Volume in µl | Expected size in bp |
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 12 | |
| 1 | - | ||
| 2 | PCR of pSB1A3+CFP | 6 | ~1000 |
| 3 | PCR of pSB1A3+YFP | 6 | ~1000 |
| 4 | PCR of pSB1K3+CFP | 6 | ~1000 |
| 5 | PCR of pSB1K3+YFP | 6 | ~1000 |
Results:
- according to PCR all 4 plasmids carry the insert
DNA extraction of mVenus and Cerulean
Investigators: Nicole
Time: 2011-08-16, 15:00-16:00
Aim: DNA of reporter genes mVenus and Cerulean
Materials
- Agarose gel electrophoresis of mVenus and Cerulean, done by Steffi previously
- Machery-Nagel NucleoSpin Extract II, protocol for DNA extraction from agarose gels
Method:
- extraction done based on manufacturer's protocol
Results:
DNA concentrations measured by Nanodrop
- cmVenus = 6.9 ng/ µl
- cCerulean = 10.6 ng/ µl
Agarose gel electrophoresis test digested pPDV100 (strategy 1)
Investigators: Sabine
Aim: control of created pPDV100
Time: 2011-08-16,10:00-12:00
Materials and Method:
- 0,75 % agarose gel</b>
- 100 V
- 1 h
expected bands:
- SfiI digestion pak bla KDIR with mdnA insert: 200 bp and 4,3 kb
- SfiI digetion pak bla KDIR without mdnA insert: 800 bp and 4,3 kb
- PvuII digestion pak bla KDIR with mdnA insert: 100 bp, 1860 bp and 2550 bp
- PvuII digestion pak bla KDIR with mdnA insert: 100 bp, 2480 bp and 2550 bp
Results:
- clone 2 may be a positive clone
Further tasks:
- send pPDV of clone 2 to Eurofins for sequencing
Sequencing of pPDV100 of clone 2 (strategy 1)
Investigators: Sabine
Aim: Control of created pPDV100
Time: 2011-08-16,13:00-14:00
Material/Method:
- Miniprep of clone 2
- Sequencing Primer: sf_mdna_1
- Freelabels for Value ReadTube (MWG Eurofins)
- DNA concentration 70 ng/ µl in a total volume of 15 µl
- Primer concentration: 15 pmol/µl in the total volume of 15 µl (mix)
Further tasks:
- Analyzing sequencing results
Digestion of pSB1A3/pSB1K3 with CFP/YFP and PCR fragments of the promoters (Ara, Lac, constitutive)
Time: 2011-08-16
Investigators: Nicole, Niels, Jessica
Aim:
Preparation for ligation of promoters into pSB1A3, pSB1K3 carrying CFP/YFP
Materials:
- minipreps of:
- pSB1A3+YFP I clone III: 179.2 ng/µl(from 2011-08-15)
- pSB1A3+CFP, pSB1K3+CFP, pSB1K3+YFP from 2011-08-05
- purified PCR fragments (Ara, Lac, constitutive) from 2011-08-02
- EcoRI-HF, XbaI
- Buffer 4
- 100x BSA
Method:
Digestion protocol (following iGEM distribution protocol for linearized backbones):
- Mastermix
- 2 µl NEB Buffer 4
- 0.2 µl BSA
- 1 µl EcoRI-HF
- 1 µl PstI-HF
- 10,8 µl pure water
- total: 15 µl
- reaction mix:
- 15 µl mastermix + 5 µl DNA
- Incubation:
- 37°C for 1 h
- Heat deactivation:
- 80°C for 20 min
Further Task:
- gel electrophoresis
- gel extraction and purification
 "
"