Team:ETH Zurich/Biology/Detector
From 2011.igem.org
(→Where it comes from...) |
|||
| Line 43: | Line 43: | ||
== Where it comes from... == | == Where it comes from... == | ||
| - | The xylR gene in ''Pseudomonas putida'' encode for a 566 aa long regulator protein that activates the degradation of toluene and xylenes. In ''P. putida'' the complete upper Tol pathway is under control of the toluene-responsive promoter P<sub>U</sub>, in the presents of xylene XylR binds to P<sub>U</sub> and activates it [[#Ref5|[5]]] The five genes xylC-xylM-xylA-xylB-xylN are expressed, xylC encodes for the benzaldehyde dehydrogenase, xylM and xylA for subunits of the xylene oxygenase and xylB for benzyl alcohol dehydrogenase [[#Ref6|[6]]]. XlyN is not required for the degradation of xylene but its part of the whole transcription unit. The DNA binding domain of XylR binds to a 40 bp long upstream | + | The ''xylR'' gene in ''Pseudomonas putida'' encode for a 566 aa long regulator protein that activates the degradation of toluene and xylenes. In ''P. putida'' the complete upper Tol pathway is under control of the toluene-responsive promoter P<sub>U</sub>, in the presents of xylene XylR binds to P<sub>U</sub> and activates it [[#Ref5|[5]]] The five genes ''xylC-xylM-xylA-xylB-xylN'' are expressed, ''xylC'' encodes for the benzaldehyde dehydrogenase, ''xylM'' and ''xylA'' for subunits of the xylene oxygenase and ''xylB'' for benzyl alcohol dehydrogenase [[#Ref6|[6]]]. XlyN is not required for the degradation of xylene but its part of the whole transcription unit. The DNA binding domain of XylR binds to a 40 bp long upstream activating sequence (USA) located 150 bp upstream from the transcription start site of the σ<sup>54</sup> P<sub>U</sub>. DNA looping between the USA and the -12/-24 motifs of the σ<sup>54</sup>-RNAP facilitates the activation of the holoenzyme by XylR. |
| + | |||
| + | |||
| + | XylR itself represses its own production. | ||
== How we used it == | == How we used it == | ||
Revision as of 08:34, 21 September 2011
| Sensors |
| ||
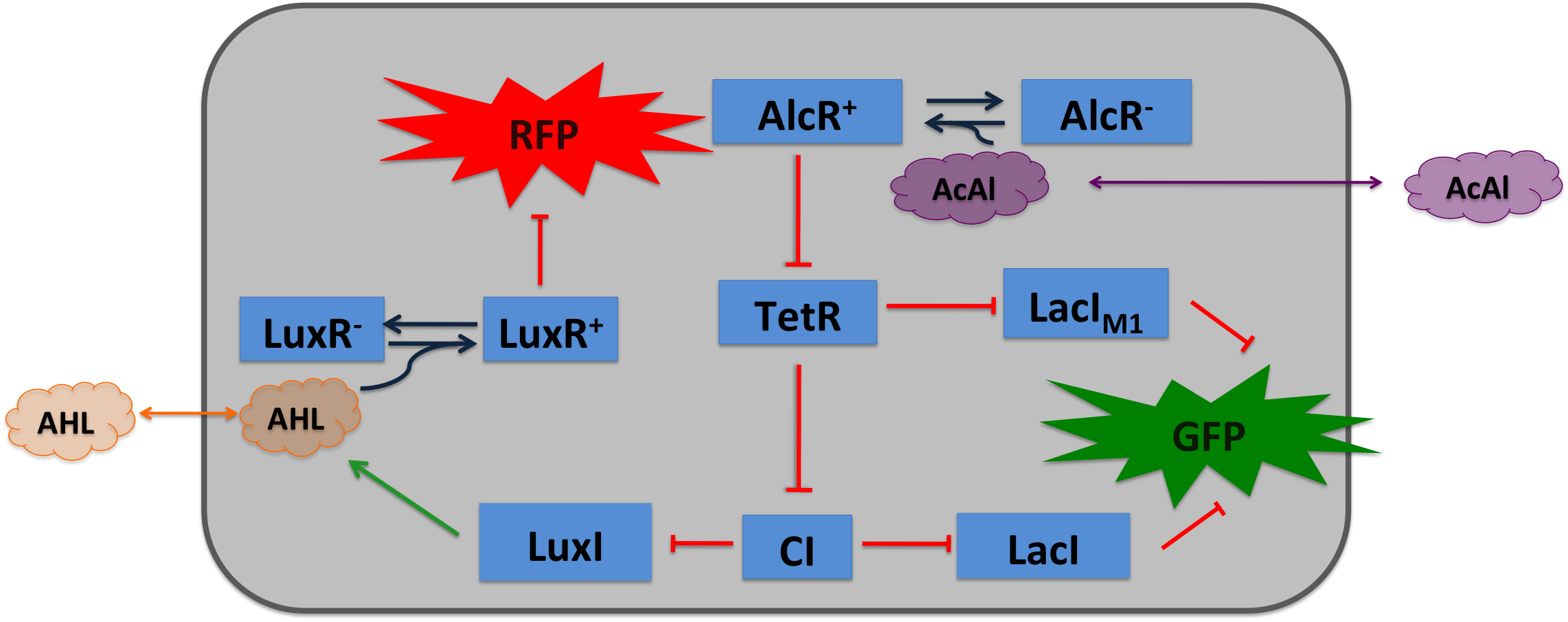
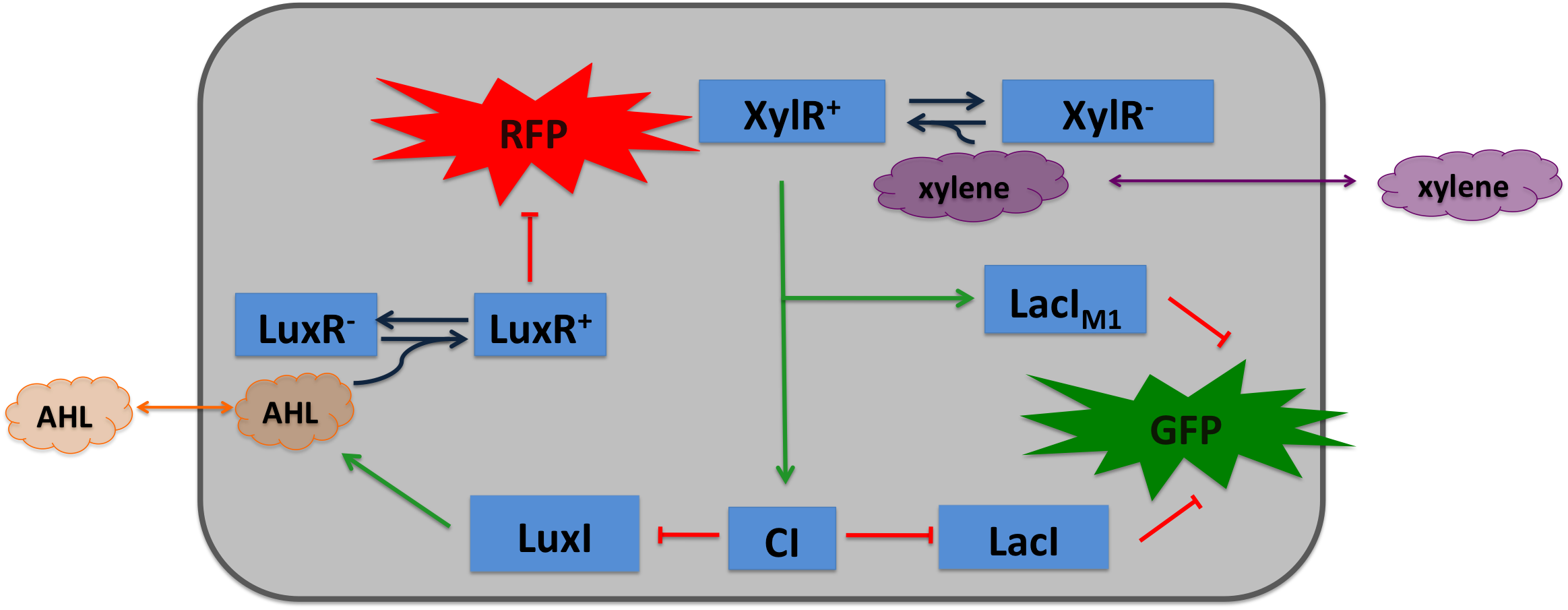
| Cigarette smoke contains a lot of different toxic and carcinogenic components. In biology a lot of sensors for such components exist mostly to induce their degradation. One is the acetaldeyhde system in aspergillus nidulans, the activator AlcR binds to it operator site if acetaldeyhde is present [1]. An other one is the xylene sensing system in Pseudomonas putida [2] and the one for styrene in Pseudomonas sp. [3], both also work as activators. | |||
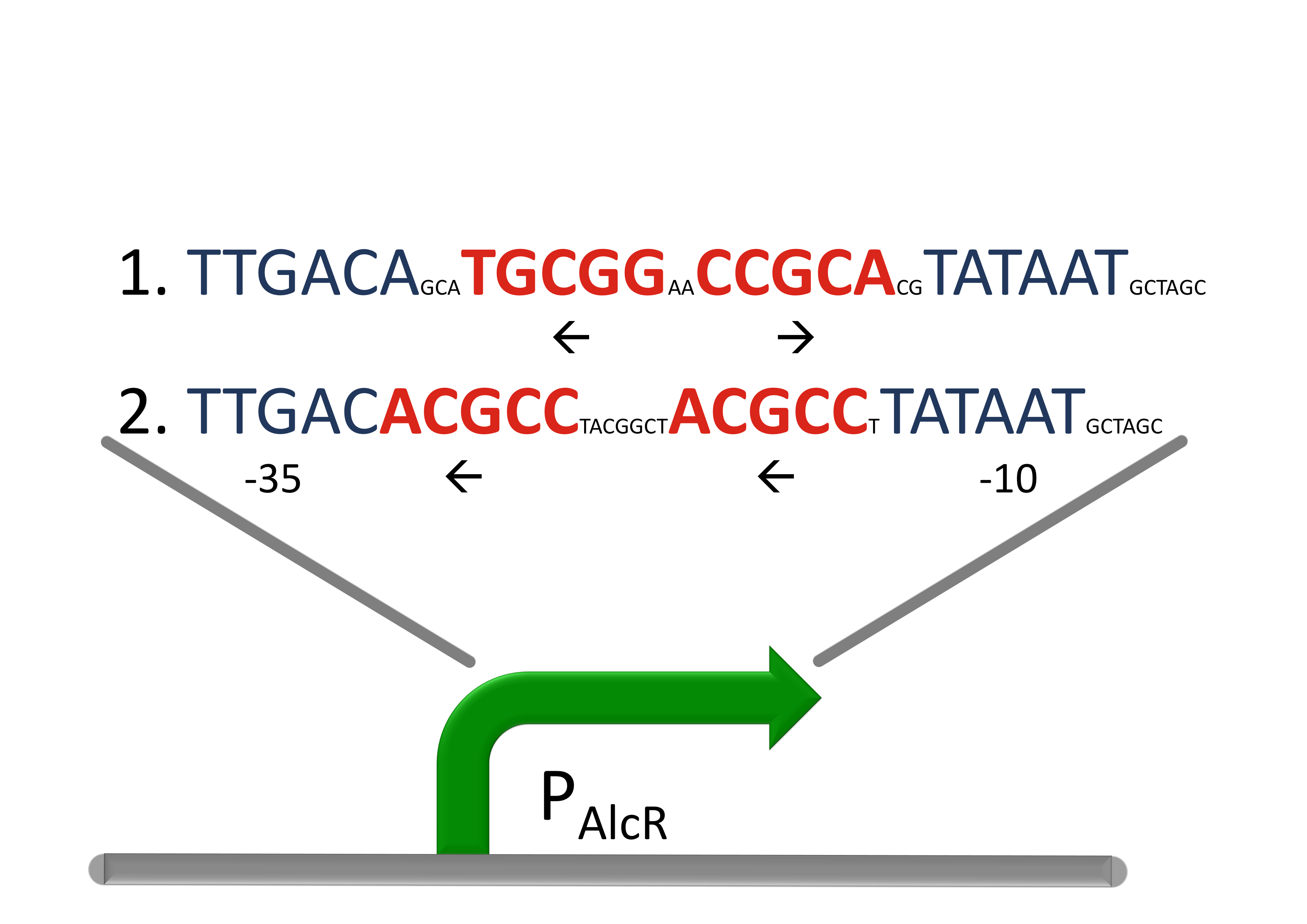
Acetaldehyde SensorWhere it comes from...In aspergillus nidulans the alcR gene encodes a regulator protein which induces the expression of the ethanol utilization (alc) pathway if the co-inducer acetaldehyde is present. Ethanol is converted to acetaldehyde by the alcohol dehydrogenase, acetaldehyde is afterward metabolized to acetate by the aldehyde dehydrogenase. Both genes alcA and aldA are regulated by AlcR. AlcR also regulate its one expression by autoactivation [1]. The AlcR DNA binding domain contains a zinc binuclear cluster, which can bind either to symmetric and assymetric sites with same affinity. It binds as a monomer, but 2 molecules can bind to inverted repeats in a noncoperative manner. For high affinity binding additional DNA sequences upstream the zinc cluster were identified [4]. How we used itBecause funghi activator normally does not work in E. coli, we modified AlcR in a way that it acts as a repressor. The mechanism of how AlcR activates transcription is not completely known what makes the process of redesigning it very challenging. A second challenge is the different codon usage in apergillus nidulans and E.coli. The Codon adaptation index (CAI) of aspergillus nidulans AlcR is 0.7 in E. coli, especially at the beginning of the gene a few very rare codons were included. To get a fully E. coli-optimized gene, we ordered the gene codon optimized. To get a partly codon optimized version, we exchanged the rare codons at the beginning of the aspergillus nidulans protein with PCR. For the promoter PAlcR the operator site of AlcR from apergillus nidulans was placed between the -10 and -35 region of the bacterial promoter. As DNA targets we used two different versions, one with direct-repeat targets and one with inverted [1]. The idea is that in present of acetaldehyde AlcR will bind to the operon and block the binding of the RNA-polymerase. This only works if the mechanism of AlcR binding does not involves conformational changes or something similar. The AlcR promotor is cloned in front of the TetR repressor to introduce the bandpass.
Acetaldehyde is naturally degraded by E. coli generating a concentration gradient in our tube. At the position with the required concentration of acetaldehyde for the bandpass-filter GFP is expressed, at all other position it is repressed, resulting in a single GFP band. By tuning the input of acetaldehyde in the tube the GFP band moves. If the acetaldehyde concentration reaches a certain very high level the whole tube expresses GFP. |
Xylene SensorWhere it comes from...The xylR gene in Pseudomonas putida encode for a 566 aa long regulator protein that activates the degradation of toluene and xylenes. In P. putida the complete upper Tol pathway is under control of the toluene-responsive promoter PU, in the presents of xylene XylR binds to PU and activates it [5] The five genes xylC-xylM-xylA-xylB-xylN are expressed, xylC encodes for the benzaldehyde dehydrogenase, xylM and xylA for subunits of the xylene oxygenase and xylB for benzyl alcohol dehydrogenase [6]. XlyN is not required for the degradation of xylene but its part of the whole transcription unit. The DNA binding domain of XylR binds to a 40 bp long upstream activating sequence (USA) located 150 bp upstream from the transcription start site of the σ54 PU. DNA looping between the USA and the -12/-24 motifs of the σ54-RNAP facilitates the activation of the holoenzyme by XylR.
How we used itXylene is not degraded naturally by E. coli. To generate a concentration gradient of xylene we engineered its degradation in SmoColi by included the upper Tol pathway. The activator XylR is known to work in E.coli as in Pseudomonas putida. In this second case we used XylR as a positive input for our bandpassfilter.
|
References[1] [http://www.ncbi.nlm.nih.gov/pubmed/2834622 R. Locklngton, C. Scazzocchio, D. Sequeval, M. Mathieu, B. Felenbok: Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans, Mol Microbiol., 1987, 1: 275-81] [2] [http://aem.asm.org/cgi/content/abstract/64/2/748 Sven Panke, Juan M. Sánchez-Romero, and Víctor de Lorenzo: Engineering of Quasi-Natural Pseudomonas putida Strains for Toluene Metabolism through an ortho-Cleavage Degradation Pathway, Appl Environ Microbiol, 1998, 64: 748-751] [3] [http://aem.asm.org/cgi/content/abstract/64/6/2032 Sven Panke, Bernard Witholt, Andreas Schmid, and Marcel G. Wubbolts: Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120, Appl Environ Microbiol, 1998, 64: 2032-2043] [4] [http://www.ncbi.nlm.nih.gov/pubmed/9182587 Lenouvel F, Nikolaev I, Felenbok B.:In vitro recognition of specific DNA targets by AlcR, a zinc binuclear cluster activator different from the other proteins of this class, J Biol Chem. 1997, 272(24):15521-6.] [5] [http://www.springerlink.com/content/r65l105rg21t1l73/ Manabu Gomada, Sachiye Inouye, Hiromasa Imaishi, Atsushi Nakazawa and Teruko Nakazawa: Analysis of an upstream regulatory sequence required for activation of the regulatory gene xylS in xylene metabolism directed by the TOL plasmid of Pseudomonas putida, Molecular and General Genetics MGG, 1992, 233:419-426] [6] [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210316/ S Harayama, M Rekik, M Wubbolts, K Rose, R A Leppik, and K N Timmis: Characterization of five genes in the upper-pathway operon of TOL plasmid pWW0 from Pseudomonas putida and identification of the gene products, 1989,171(9): 5048–5055]
|
 "
"