Team:Wageningen UR/Project/CompleteProject1Description
From 2011.igem.org
RubenvanHeck (Talk | contribs) |
m (→Synchroscillator) |
||
| Line 30: | Line 30: | ||
====1. Introduction==== | ====1. Introduction==== | ||
| - | The aim of this project is to design and implement a system exhibiting sustained oscillatory protein expression which is synchronized across a population of spatially constrained E. coli cells. The principles that govern this type of behaviour have been studied both in theory and in practice, and as such there exists a solid foundation to apply these ideas in the context of the iGEM competition. In essence, this project consists of constructing a plasmid which contains genes encoding proteins which reciprocally affect each other’s expression in a reliable manner, and experimentally measuring the expression dynamics to test the predictive value of a mathematical model. Due to the specificity of the required system parameters, and resulting difficulty in experimentally verifying the phenomena we wished to observe, special considerations regarding the experimental set-up had to be made. We hope that this system might be employed as a pace-making device to drive more complex genetic circuits requiring time-dependent gene expression, or as a component in sophisticated metabolic engineering applications. | + | The aim of this project is to design and implement a system exhibiting sustained oscillatory protein expression which is synchronized across a population of spatially constrained ''E. coli'' cells. The principles that govern this type of behaviour have been studied both in theory and in practice, and as such there exists a solid foundation to apply these ideas in the context of the iGEM competition. In essence, this project consists of constructing a plasmid which contains genes encoding proteins which reciprocally affect each other’s expression in a reliable manner, and experimentally measuring the expression dynamics to test the predictive value of a mathematical model. Due to the specificity of the required system parameters, and resulting difficulty in experimentally verifying the phenomena we wished to observe, special considerations regarding the experimental set-up had to be made. We hope that this system might be employed as a pace-making device to drive more complex genetic circuits requiring time-dependent gene expression, or as a component in sophisticated metabolic engineering applications. |
| Line 40: | Line 40: | ||
====2. Mechanism==== | ====2. Mechanism==== | ||
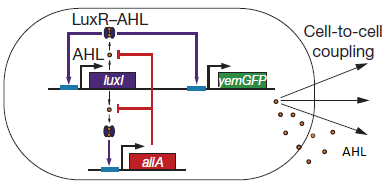
| - | There are a number of genetic circuit topologies that have the potential to exhibit oscillatory behaviour under the right conditions. However, the requirement that the oscillations be synchronized posed a constraint on the components that could be used. The starting point for our genetic circuitry was a design recently published in the article “A synchronized quorum of genetic clocks” by Danino et al. This design combines elements of the Vibrio fischeri quorum sensing system with a quorum quenching enzyme from Bacillus subtilis, resulting in coupled positive and negative feedback loops which regulate the expression of a reporter protein. | + | There are a number of genetic circuit topologies that have the potential to exhibit oscillatory behaviour under the right conditions. However, the requirement that the oscillations be synchronized posed a constraint on the components that could be used. The starting point for our genetic circuitry was a design recently published in the article “A synchronized quorum of genetic clocks” by Danino et al. This design combines elements of the ''Vibrio fischeri'' quorum sensing system with a quorum quenching enzyme from ''Bacillus subtilis'', resulting in coupled positive and negative feedback loops which regulate the expression of a reporter protein. |
| Line 53: | Line 53: | ||
The enzyme '''LuxI''' is an acyl-homoserine-lactone synthase which produces the intercellular signalling molecule N-(3-oxohexanoyl)-homoserine lactone. Placing LuxI under control of the pRight promoter results in a positive feedback loop: when increases in cell density cause the intracellular AHL concentration to rise above the activation threshold of the pRight promoter, the transcription rate of the LuxI gene is increased which in turn results in the production of more AHL. | The enzyme '''LuxI''' is an acyl-homoserine-lactone synthase which produces the intercellular signalling molecule N-(3-oxohexanoyl)-homoserine lactone. Placing LuxI under control of the pRight promoter results in a positive feedback loop: when increases in cell density cause the intracellular AHL concentration to rise above the activation threshold of the pRight promoter, the transcription rate of the LuxI gene is increased which in turn results in the production of more AHL. | ||
| - | '''AiiA''' is an enzyme from B. subtilis which degrades AHL. Its biological function is to interfere with the quorum sensing signals of other bacteria. Placing it under control of the pRight promoter results in negative feedback as a response to increasing AHL concentrations. | + | '''AiiA''' is an enzyme from ''B. subtilis'' which degrades AHL. Its biological function is to interfere with the quorum sensing signals of other bacteria. Placing it under control of the pRight promoter results in negative feedback as a response to increasing AHL concentrations. |
The reporter molecule Green Fluorescent Protein ('''GFP''')is also regulated by the pRight promoter and provides a quantitative (albeit delayed) indication of the AHL concentration the cell is exposed to at a given point in time. | The reporter molecule Green Fluorescent Protein ('''GFP''')is also regulated by the pRight promoter and provides a quantitative (albeit delayed) indication of the AHL concentration the cell is exposed to at a given point in time. | ||
| - | LuxI, AiiA and GFP are all tagged for rapid degradation. Due to differences in the synthesis and degradation rates of LuxI and AiiA, there exists a space of conditions within which periodic oscillations in AHL concentration, and concomitant | + | LuxI, AiiA and GFP are all tagged for rapid degradation. Due to differences in the synthesis and degradation rates of LuxI and AiiA, there exists a space of conditions within which periodic oscillations in AHL concentration, and concomitant oscillatory protein expression can emerge. Under most conditions, the level of AHL within a population of cells will quickly reach a steady state. However, by simulating the system using a quantitative biochemical model, it is possible to predict conditions under which oscillations are likely to occur. See [https://2011.igem.org/Team:Wageningen_UR/Project/ModelingProj1 Modeling page] for details. |
=====Designs===== | =====Designs===== | ||
| Line 64: | Line 64: | ||
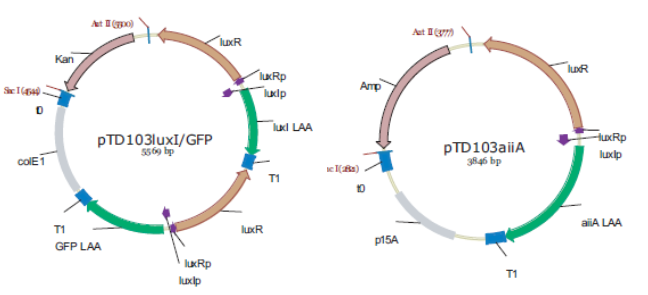
'''Fig.4.''' ''Synchronized Oscillator plasmids using natural quorum sensing system (Danino et al. 2010)'' | '''Fig.4.''' ''Synchronized Oscillator plasmids using natural quorum sensing system (Danino et al. 2010)'' | ||
| - | The first design we implemented was a BioBrick part based reconstruction of the plasmids used by Danino et al. We intended to make as accurate a replica as possible in order to confirm the previously published results, and to test the viability of our experimental [https://2011.igem.org/Team:Wageningen_UR/Project/Devices platform]. However, there are a few differences in between the original Hasty system and our replica. While the Hasty system employs the natural lux promoter which contains divergent pLeft and pRight elements (Fig 4), the BioBrick parts we employed have both elements in the same orientation. Both the original and our system contain | + | The first design we implemented was a BioBrick part based reconstruction of the plasmids used by Danino et al. We intended to make as accurate a replica as possible in order to confirm the previously published results, and to test the viability of our experimental [https://2011.igem.org/Team:Wageningen_UR/Project/Devices platform]. However, there are a few differences in between the original Hasty system and our replica. While the Hasty system employs the natural lux promoter which contains divergent pLeft and pRight elements (Fig 4), the BioBrick parts we employed have both elements in the same orientation. Both the original and our system contain three copies of the luxR gene under control of the pLeft element. Furthermore, a different (high copy) backbone was used during the functional validation of the parts, as opposed to the low copy backbone employed by Hasty. |
[[File:Hasty1.png|x67px|link=http://partsregistry.org/Part:BBa_K546005]] | [[File:Hasty1.png|x67px|link=http://partsregistry.org/Part:BBa_K546005]] | ||
| Line 79: | Line 79: | ||
'''Double Tunable Synchronized Oscillator''' | '''Double Tunable Synchronized Oscillator''' | ||
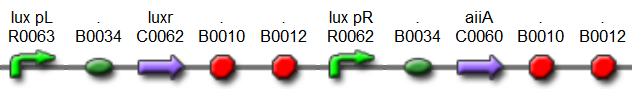
| - | The aforementioned constraints are due to the fact that there are no means by which the expression kinetics of LuxI and AiiA can be externally influenced. In order to gain more control over the expression dynamics, we refactored the system and introduced “tuning knobs” in the form of chemically inducible hybrid promoters (pR/LacI inducible by IPTG and pR/tetR inducible by anhydrotetracycline). | + | The aforementioned constraints are due to the fact that there are no means by which the expression kinetics of LuxI and AiiA can be externally influenced. In order to gain more control over the expression dynamics, we refactored the system and introduced “tuning knobs” in the form of chemically inducible hybrid promoters (pR/LacI inducible by IPTG and pR/tetR inducible by anhydrotetracycline). The addition of two independent control variables substantially expands the parameter space (cell density, flow rate) within which oscillations can occur. We also made the design more streamlined by taking advantage of the great diversity of available BioBrick quorum sensing parts in order to remove redundant elements and fit the entire system onto a single plasmid (a 30% reduction in size compared to the original design). Note: in order take advantage of the tuning capabilities it is necessary to co-transform a plasmid which constitutively expresses LacI or TetR, or both. |
[[File:Dt.png|x67px|link=http://partsregistry.org/Part:BBa_K546546]] | [[File:Dt.png|x67px|link=http://partsregistry.org/Part:BBa_K546546]] | ||
Revision as of 21:47, 20 September 2011
 "
"