Team:LMU-Munich/Project/Description
From 2011.igem.org
(→Primer sequences) |
|||
| Line 95: | Line 95: | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<!-- Include the next line at the end of every page --> | <!-- Include the next line at the end of every page --> | ||
{{:Team:LMU-Munich/Templates/Page Footer}} | {{:Team:LMU-Munich/Templates/Page Footer}} | ||
Revision as of 13:50, 18 September 2011
Overall project
Metals and especially heavy metals are highly prescribed in concentrations in the drinking water ordinance. Qualifying and quantifying these by standard chemical methods is costly and complicated.
Bacteria sense metals in their surrounding in order to change their expression profile or react in order to adapt and accomodate to their environment.
Using these sensors from (mostly) bacteria we create biosensors by linking them to the expression of a reporter (e.g. green glowing by the green fluorescent protein GFP). To not only qualify but also to quantify the metals, it is also necessary to measure the output by given input (metal concentration) for each of these biosensors. Afterwards one can determine the metal concentration by measuring the output.
The quantification needs heavy high-tech machinery ... something not always given ... especially in free field. So a qualification of metals with an easy-to-see output is also needed.
In the end our team hopes to have not only a set of metallsensors for precise quantification of a group of (heavy) metalls, but also an outdoor kit for qualifying metalls in more remote areas. With these it might be more easy and cheaper to determine the content of metals in our drinking water.
Project Details
This year’s project by the iGEM-team from Ludwig-Maximilians-University in Munich uses natural biosensors to detect the concentration of different metals. We have the vision to develop a set of bacterial metal sensors for easy qualitative and quantitative measurement of toxic metals just by reading the output after adding the water test sample.
We use two different kinds of metal sensors. The ones in the first category work with reporter genes that lay downstream of an inducible promoter. The respective promoter is activated or deactivated by a specific metal-sensitive protein which binds to DNA dependent on the presence of that metal. As a consequence of this statistical event, there is a concentration-dependent transcription of the reporter gene, which is either GFP, luxAB or lacZ´.
The second kind of metal sensors directly uses the characteristics of special proteins to obtain a measurement of the metal, e.g. by analyzing the activity of an enzyme that needs a special metal ion as a cofactor.
For easy handling and compatibility we use the E. coli strain DH5α.
Reporter genes
Promoter-based Sensors
All genes are transcriptionally fused to the reporter gene, which means that approximately 50 nucleotides from the originally transcribed gene still remain with the promoter to ensure correct read-off. Downstream of this short open reading frame, the reporter gene with its own ribosome binding site is added.
source(s):
- [http://www.ncbi.nlm.nih.gov/pubmed?term=coordinating%20intracellular%20chivers Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors] Iwig JS, Chivers PT
- [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1236639/?tool=pubmed Complex transcriptional control links NikABCDE-dependent nickel transport with hydrogenase expression in Escherichia coli] RoweJS, Starnes GL, Chivers PT
- [http://www.ncbi.nlm.nih.gov/pubmed/10787413 Regulation of high affinity nickel uptake in bacteria] Ni2+-Dependent interaction of NikR with wild-type and mutant operator sites. Chivers PT, Sauer RT
- [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC93426/?tool=pubmed Isolation and characterization of the nikR gene encoding a nickel-responsive regulator in Escherichia coli] de Pina K, Desjardin V, Mandrand-Berthelot MA, Giordano G, Wu LF
source(s):
- [http://onlinelibrary.wiley.com/doi/10.1111/j.1365-2958.2006.05369.x/pdf Nickel homeostasis in Escherichia coli – the rcnR-rcnA efflux pathway and its linkage to NikR function] Iwig JS, Rowe JL, Chivers PT
- [http://www.ncbi.nlm.nih.gov/pubmed?term=coordinating%20intracellular%20chivers Coordinating intracellular nickel-metal-site structure-function relationships and the NikR and RcnR repressors] Iwig JS, Chivers PT
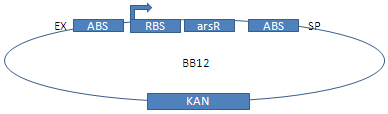
Since the regulator arsR does not exist in E. coli, it is coded on our plasmids. The pars-promoter with two binding sites has been multiplied via PCR with Phusion polymerase and primers E and F at an annealing temperature of 50°C. arsR with a constantly active promoter was cloned from BBa_K3562 and fused using 3A-assembly.
source(s):
Einzelne Systeme erklären
Bilder!!!
Bild Wellplatte mit bunten Farben
The Experiments
working BioBrick erläutern
Results
welche BioBricks sind fertig und funktionieren hier reinschreiben!!!
 "
"