Team:Freiburg/Description
From 2011.igem.org
(Difference between revisions)
(→Precipitator) |
(→Precipitator) |
||
| Line 4: | Line 4: | ||
[[File:Freiburg11_precip1.png|right|thumb|caption|200px|Molecular model of the Precipitator]] | [[File:Freiburg11_precip1.png|right|thumb|caption|200px|Molecular model of the Precipitator]] | ||
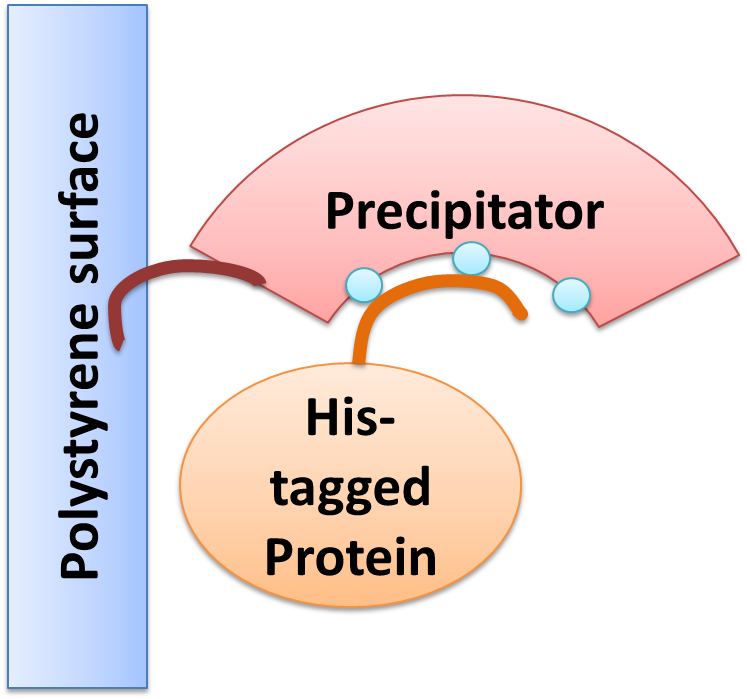
| - | We created a cellular, self-replicating purification device for His-tagged proteins. It is a completely artificial fusion protein, which consists of a repeating LRRNT motif domain, coordinating Ni2+ Ions on its surface capped on N and C terminal ends by a hagfish sequence of a similar LRRNT motif. A second domain, a short hydrophobic peptide stretch, binds a polystyrene surface | + | ===The Concept=== |
| - | After expression of the Precipitator in a light inducible E. coli strain, the cells are lysed and the lysate is taken up | + | We created a cellular, self-replicating purification device for His-tagged proteins. It is a completely artificial fusion protein, which consists of a repeating LRRNT motif domain, coordinating Ni2+ Ions on its surface capped on N and C terminal ends by a hagfish sequence of a similar LRRNT motif. We named this construct “THE PRECIPITATOR”. A second domain, a short hydrophobic peptide stretch, binds a polystyrene surface, called the plastic binding domain. |
| - | The underlying mechanism is comparable to Ni-NTA columns. Our Precipitator protein binds on the surface of the pipette, presenting the chelated Nickel ions. Free coordination sites of the Nickel ions are exposed, so that a His-tagged protein can attach to them. Cells expressing a His-tagged protein can be dissolved by the heat inducible lysis device. Subsequently, when the lysate is taken up with a serological pipette coated with the Precipitator protein, the His-tagged proteins bind to it. Cell debris is then washed off, while the His-tagged protein stays and is eluted afterwards, in the same fashion as done in Ni-NTA columns with imidazole solutions, increasing in concentration. The His-tagged protein is finally captured in a distinct fraction. | + | After expression of the Precipitator in a light inducible E. coli strain, the cells are lysed and the lysate is taken up with a serological pipette, in preparation of the actual protein purification steps. |
| + | The underlying mechanism is comparable to Ni-NTA columns. Our Precipitator protein binds on the surface of the pipette, presenting the chelated Nickel ions. Free coordination sites of the Nickel ions are exposed, so that a His-tagged protein can attach to them. Cells expressing a His-tagged protein can be dissolved by the heat inducible lysis-device. Subsequently, when the lysate is taken up with a serological pipette coated with the Precipitator protein, the His-tagged proteins bind to it. Cell debris is then washed off, while the His-tagged protein stays and is eluted afterwards, in the same fashion as done in Ni-NTA columns with imidazole solutions, increasing in concentration. The His-tagged protein is finally captured in a distinct fraction. | ||
| + | |||
| + | ===Part design=== | ||
| + | The Precipitator BBa_K608406 protein is made of an artificial Leucine Rich Repeat (LRR) as the middle part of our own design, capped by C and N-terminal hagfish domain fragments.. This part is one version of three different designed to bind nickel by histidines, grouped together pointing away from the horseshoe shaped protein. Please see modeling for more details | ||
| + | |||
| + | Bacterial LRR Consensus of the central LRR fragment: | ||
| + | LxxLxLxxNxLxxLPxxLPxx | ||
| + | |||
| + | Protein sequence: <br> | ||
| + | <span style="color:violet;"> | ||
| + | CPSRCSCSGTEIRCNSKGLTSVPTGIPSS <br> | ||
| + | ATRLELESNKLQSLPHGVFDK </span><br><span style="color:green;"> | ||
| + | LTQLTKSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br> | ||
| + | LEVLDVSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br> | ||
| + | LEVLDVSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br> | ||
| + | LEVLDVSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br> | ||
| + | LEVLDVSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br> | ||
| + | LEVLDVSNN<span style="color:red;">H</span>L<span style="color:red;">H</span>SLPDNLPAS <br></span><span style="color:gold;"> | ||
| + | LKELALDTNQLKSVPDGIFDR <br> | ||
| + | LTSLQKIWLHTNPWDCSCPRIDY <br> | ||
| + | LSRWLNKNSQKEQGSAKCSGSGKPVRSIICP <br></span> | ||
| + | |||
| + | This protein can be used to complex Nickel or Cobalt. Histidines are positioned in such a way, that they can coordinate the ions from two to four orthogonal oriented directions. Free binding sites of the ions are then exposed, so that a His-tagged protein can attach to them. This protein can be used to complex up to 4 Nickel or Cobalt. | ||
| + | The underying design of the protein is of a particular interest, too. LRR are highly conserved motifs throughout evolution. They appear in all kingdoms of life in almost every thinkable role (Ligases, Receptors, Toxins etc.). Their core is highly conserved and provides a very stable backbone, while the non-conserved aminoacids are almost freely interchangeable. | ||
| + | Here we investigated an optimal set of non-conserved aminoacids by analysing large sets of similar proteins and databases. You can use this piece of work as a template to design your own protein and give it any function you like, by simply interchanging aminoacids and fusing other domains on the N or C termini. To guarantee proper folding and to shield off the hydrophobic core, a well studied fragment of an LRR protein coming from hagfish was used. This efficiency of this technique was proven before.(REFERENZ). To find out the most likely folding, we designed many different protein sequences, trying out a variety of sets of non coding aminoacids for the LRR and submitted these to the I-TASSER structure prediction | ||
| + | |||
| + | We only submitted one of the three versions, to reduce redundancy in the registry. Please contact us for any questions. | ||
==<span style="color:green;">Green light receptor</span>== | ==<span style="color:green;">Green light receptor</span>== | ||
Revision as of 11:41, 18 September 2011
 "
"
 Contact
Contact