Team:ETH Zurich/Biology/Detector
From 2011.igem.org
| Line 15: | Line 15: | ||
{| style="width:900px;background:#f0f0f0;text-align:justify;font-family: helvetica, arial, sans-serif; margin-top:25px; border-radius:10px;" cellspacing="6" cellpadding="4" | {| style="width:900px;background:#f0f0f0;text-align:justify;font-family: helvetica, arial, sans-serif; margin-top:25px; border-radius:10px;" cellspacing="6" cellpadding="4" | ||
| | | | ||
| - | == Acetaldehyde Sensor == | + | == '''Acetaldehyde Sensor''' == |
| + | |||
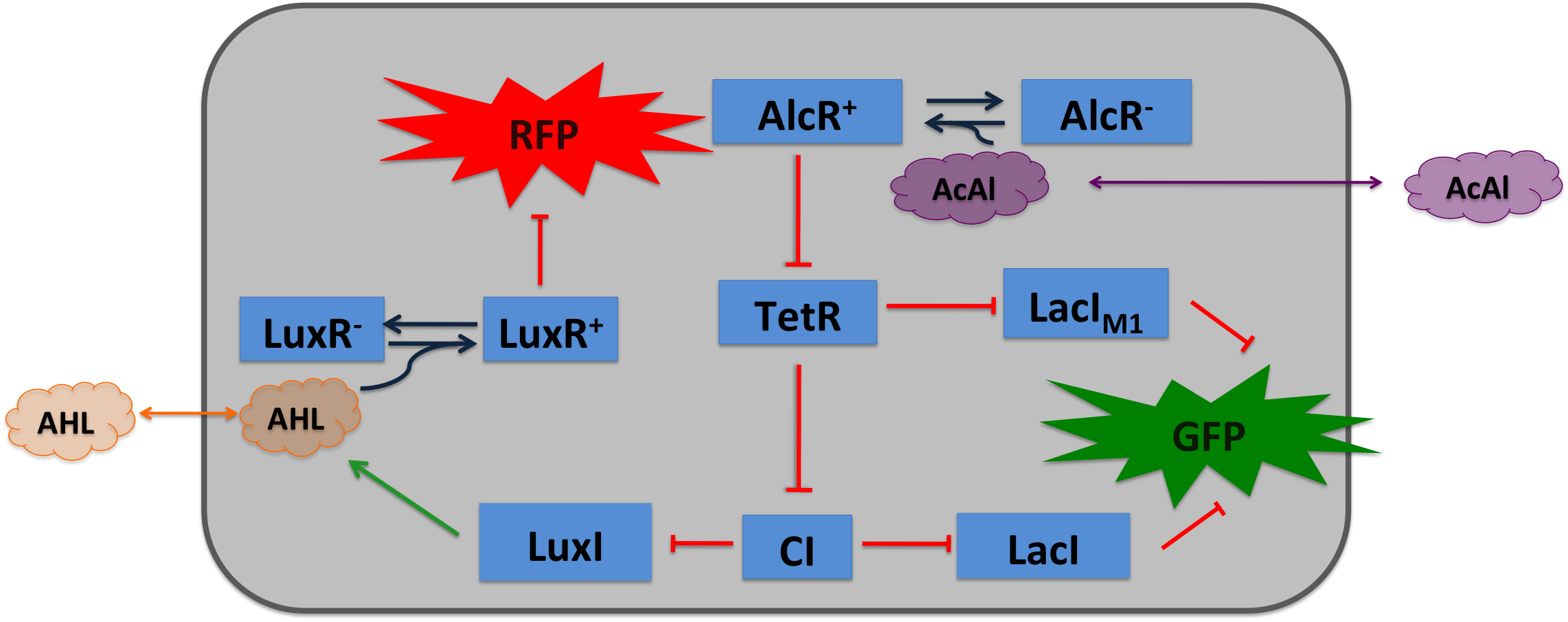
| + | Acetaldehyde is naturally degraded by ''e.coli'' generating a concentration gradient in our tube. At the position with the required concentration of acetaldehyde for the bandpass-filter GFP is expressed, at all other position it is repressed, resulting in a single GFP band. By tuning the input of acetaldehyde in the tube the GFP band moves. | ||
[[File:Design2.png|600px|center|thumb|'''Molecular mechanism of SmoColi with acetaldehyde sensor ''' ]] | [[File:Design2.png|600px|center|thumb|'''Molecular mechanism of SmoColi with acetaldehyde sensor ''' ]] | ||
| + | |||
| + | |||
| + | ==Design of a AlcR/acetaldehyde repressed promoter P<sub>AlcR</sub>== | ||
| + | |||
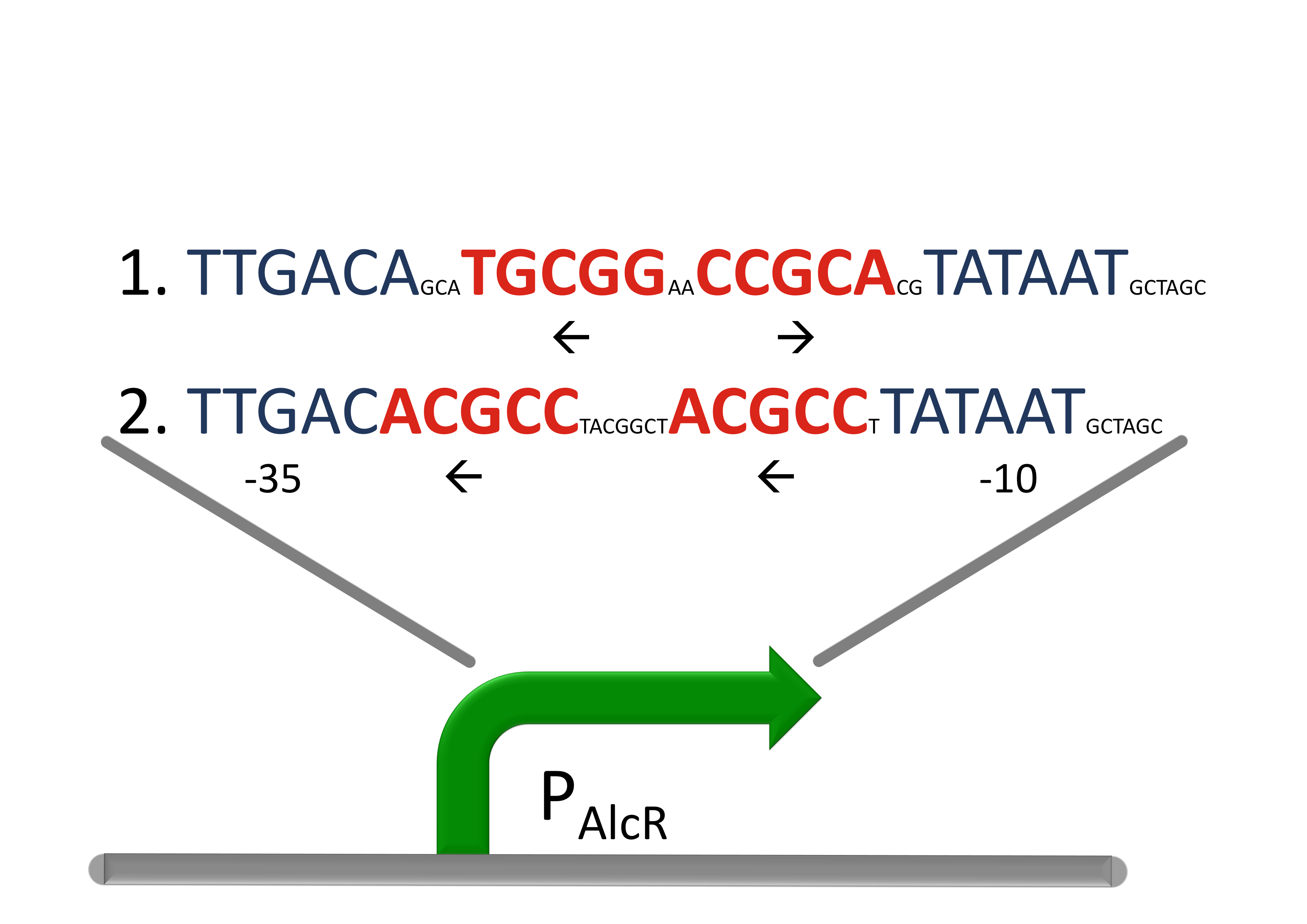
| + | Because funghi activator normally does not work in ''e.coli'', we modified AlcR in a way that it acts as a repressor. The mechanism of how AlcR activates transcription is not completely known what makes the process of redesigning it very challenging. | ||
| + | For the promoter P<sub>AlcR</sub> the operator site of AlcR from ''apergillus nidulans'' was placed between the -10 and -35 region of the bacterial promoter. As DNA targets we used two different versions, one with direct-repeat targets and one with inverted [[#Ref4|[4]]]. The idea is that in present of acetaldehyde AlcR will bind to the operon and block the binding of the RNA-polymerase. This only works if the mechanism of AlcR binding does not involves conformational changes or something similar. | ||
| + | |||
| + | [[File:AlcR_Promotordesign.png|600px|center|thumb|'''Design of the promoter for the AlcR-system in SmoColi''', the operon sequence is designed between the -10 and the -35 region (blue) of a strong promoter with inverted and direct-repeats.]] | ||
|} | |} | ||
{| style="width:900px;background:#f0f0f0;text-align:justify;font-family: helvetica, arial, sans-serif; margin-top:25px; border-radius:10px;" cellspacing="6" cellpadding="4" | {| style="width:900px;background:#f0f0f0;text-align:justify;font-family: helvetica, arial, sans-serif; margin-top:25px; border-radius:10px;" cellspacing="6" cellpadding="4" | ||
| | | | ||
| - | == Xylene Sensor == | + | == '''Xylene Sensor''' == |
| + | |||
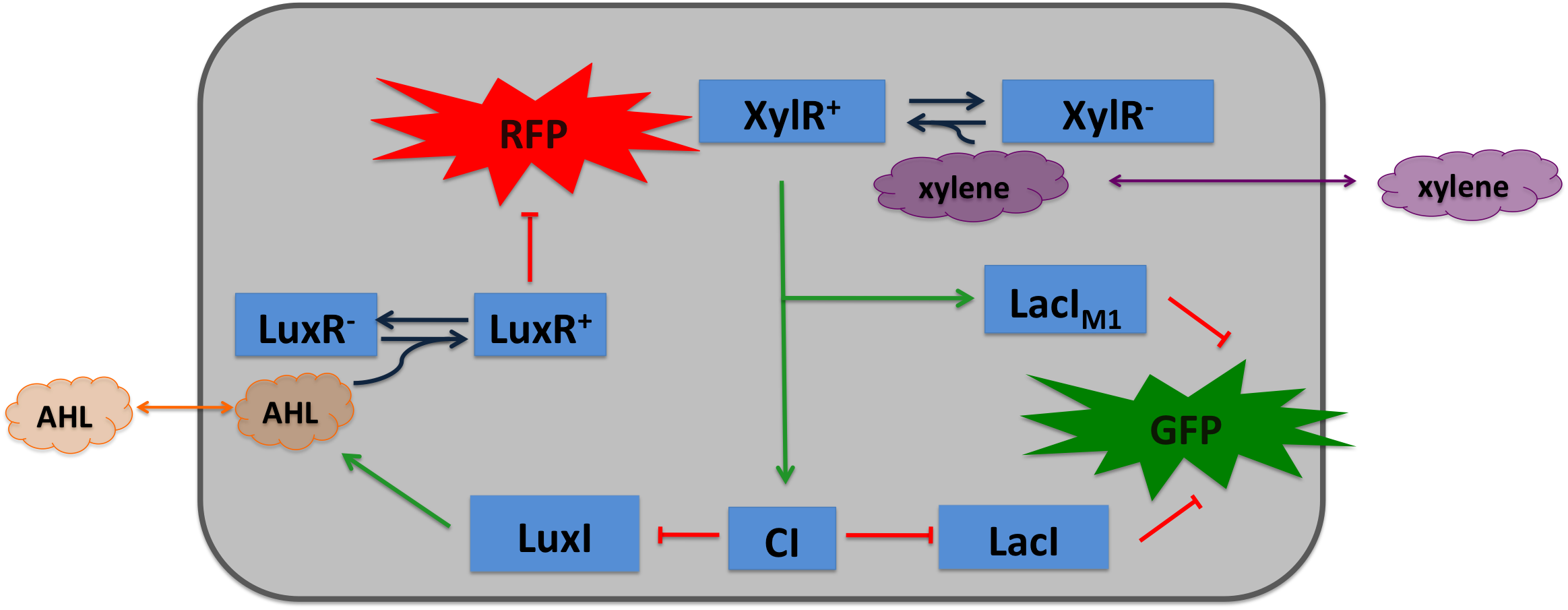
| + | Xylene is not degraded naturally by ''e.coli''. To generate a concentration gradient of xylene we engineered its degradation in SmoColi by included the upper Tol pathway. The activator XylR is known to work in ''e.coli'' as in ''Pseudomonas putida'' | ||
| + | |||
[[File:Alternative_Xylene_system.png|600px|center|thumb|'''Molecular mechanism of SmoColi with xylene sensor.''' ]] | [[File:Alternative_Xylene_system.png|600px|center|thumb|'''Molecular mechanism of SmoColi with xylene sensor.''' ]] | ||
| Line 36: | Line 49: | ||
<span id="Ref3">[3] [http://aem.asm.org/cgi/content/abstract/64/6/2032 Sven Panke, Bernard Witholt, Andreas Schmid, and Marcel G. Wubbolts: Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120, Appl Environ Microbiol, 1998, 64: 2032-2043]</span> | <span id="Ref3">[3] [http://aem.asm.org/cgi/content/abstract/64/6/2032 Sven Panke, Bernard Witholt, Andreas Schmid, and Marcel G. Wubbolts: Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120, Appl Environ Microbiol, 1998, 64: 2032-2043]</span> | ||
| + | |||
| + | <span id="Ref4">[4] [http://www.ncbi.nlm.nih.gov/pubmed/11550794 Beatrice Felenbok, Michel Flipphi and Igor Nikolaev: Ethanol Catabolism in ''Aspergillus nidulans'': A Model System for Studying Gene Regulation, Progress in Nucleic Acid Research and Molecular Biology, 69: 149-204]</span> | ||
|} | |} | ||
Revision as of 20:30, 11 September 2011
| Sensors |
| ||
| Cigarette smoke contains a lot of different toxic and carcinogenic components. In biology a lot of sensors for such components exist mostly to induce their degradation. One is the acetaldeyhde system in aspergillus nidulans, the activator AlcR binds to it operator site if acetaldeyhde is present [1]. An other one is the xylene sensing system in Pseudomonas putida [2] and the one for styrene in Pseudomonas sp. , both also work as activators [3]. | |||
Acetaldehyde SensorAcetaldehyde is naturally degraded by e.coli generating a concentration gradient in our tube. At the position with the required concentration of acetaldehyde for the bandpass-filter GFP is expressed, at all other position it is repressed, resulting in a single GFP band. By tuning the input of acetaldehyde in the tube the GFP band moves.
Design of a AlcR/acetaldehyde repressed promoter PAlcRBecause funghi activator normally does not work in e.coli, we modified AlcR in a way that it acts as a repressor. The mechanism of how AlcR activates transcription is not completely known what makes the process of redesigning it very challenging. For the promoter PAlcR the operator site of AlcR from apergillus nidulans was placed between the -10 and -35 region of the bacterial promoter. As DNA targets we used two different versions, one with direct-repeat targets and one with inverted [4]. The idea is that in present of acetaldehyde AlcR will bind to the operon and block the binding of the RNA-polymerase. This only works if the mechanism of AlcR binding does not involves conformational changes or something similar. |
Xylene SensorXylene is not degraded naturally by e.coli. To generate a concentration gradient of xylene we engineered its degradation in SmoColi by included the upper Tol pathway. The activator XylR is known to work in e.coli as in Pseudomonas putida
|
References[1] [http://www.ncbi.nlm.nih.gov/pubmed/2834622 R. Locklngton, C. Scazzocchio, D. Sequeval, M. Mathieu, B. Felenbok: Regulation of alcR, the positive regulatory gene of the ethanol utilization regulon of Aspergillus nidulans, Mol Microbiol., 1987, 1:275-81] [2] [http://aem.asm.org/cgi/content/abstract/64/2/748 Sven Panke, Juan M. Sánchez-Romero, and Víctor de Lorenzo: Engineering of Quasi-Natural Pseudomonas putida Strains for Toluene Metabolism through an ortho-Cleavage Degradation Pathway, Appl Environ Microbiol, 1998, 64: 748-751] [3] [http://aem.asm.org/cgi/content/abstract/64/6/2032 Sven Panke, Bernard Witholt, Andreas Schmid, and Marcel G. Wubbolts: Towards a Biocatalyst for (S)-Styrene Oxide Production: Characterization of the Styrene Degradation Pathway of Pseudomonas sp. Strain VLB120, Appl Environ Microbiol, 1998, 64: 2032-2043] [4] [http://www.ncbi.nlm.nih.gov/pubmed/11550794 Beatrice Felenbok, Michel Flipphi and Igor Nikolaev: Ethanol Catabolism in Aspergillus nidulans: A Model System for Studying Gene Regulation, Progress in Nucleic Acid Research and Molecular Biology, 69: 149-204] |
 "
"