Team:BU Wellesley Software/Wet Lab
From 2011.igem.org
| Line 162: | Line 162: | ||
<li> <b> Creation: It was the best of the times; it was the worst of times</b> </li> | <li> <b> Creation: It was the best of the times; it was the worst of times</b> </li> | ||
| - | Optimus Primer: More Than Meets the Eye | + | <li> <b>Optimus Primer: More Than Meets the Eye</b></li> |
After we characterized the promoters with the FACS machine and researched the TB genes using Genome Surfer, we could theoretically start building a genetic device with a TB gene fused into the fluorescent protein gene. But we still had to design a primer that can amplify the TB gene and attach it with restriction sites during a PCR reaction. This was where Optimus Primer played it major role. Because this application is integrated with Clotho, a selected sequence from the TB gene can be crosschecked with the feature list so that the amplified sequence did not contain any restriction site. By clicking the “Generate Primer”, I had this program to produce 6 primers for both the forward and reverse orientations. For each set of the 2 primers (FF and FR), Optimus Primer calculated the differences in length, the melting temperature, and the free energy. In case possible dimerization is detected, the bottom panel will turn pink. Because specific insert can be added into the 5’ and 3’ ends of the planned primer, the restriction sites E & X and S & P are added into the 2 respective ends of the primers. This step is taken because the amplified sequence needed to be combined into a genetic device for the construction of complex plasmid architecture. After the planning was completed using Optimus Primer, the provided primer sequence was ordered. The primer was then used in a PCR reaction. | After we characterized the promoters with the FACS machine and researched the TB genes using Genome Surfer, we could theoretically start building a genetic device with a TB gene fused into the fluorescent protein gene. But we still had to design a primer that can amplify the TB gene and attach it with restriction sites during a PCR reaction. This was where Optimus Primer played it major role. Because this application is integrated with Clotho, a selected sequence from the TB gene can be crosschecked with the feature list so that the amplified sequence did not contain any restriction site. By clicking the “Generate Primer”, I had this program to produce 6 primers for both the forward and reverse orientations. For each set of the 2 primers (FF and FR), Optimus Primer calculated the differences in length, the melting temperature, and the free energy. In case possible dimerization is detected, the bottom panel will turn pink. Because specific insert can be added into the 5’ and 3’ ends of the planned primer, the restriction sites E & X and S & P are added into the 2 respective ends of the primers. This step is taken because the amplified sequence needed to be combined into a genetic device for the construction of complex plasmid architecture. After the planning was completed using Optimus Primer, the provided primer sequence was ordered. The primer was then used in a PCR reaction. | ||
Revision as of 00:23, 11 September 2011
Wet Lab
Background
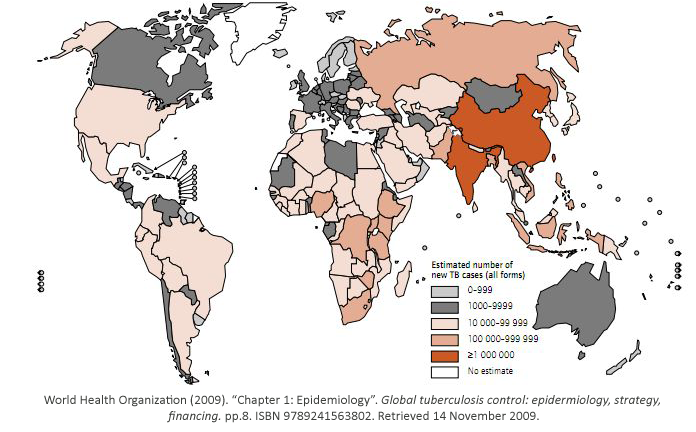
While some bacterial infections are seen as common place, others remain elusive to cure.
Tuberculosis has currently infected a third of the world’s population, and 1.7 million people
died from it in 2009. As shown, it is a worldwide problem that is prevalent in third world
countries.
Tuberculosis exists in two stages. In its latent stage, the immune system has it under
control and prevents the bacteria from reproducing. When it switches into its active stage as
a person's immune system becomes compromised, it begins to multiply and the person starts
showing symptoms of the disease.  The genetic mechanisms that trigger the change between the two
forms are not very well understood. The genetic network of tuberculosis is complicated and
studying the interactions between genes can be time consuming, requiring the construction of
many plasmids to study only one small part of the network. To look for new, faster ways of
studying this problem we turned to synthetic biology. Instead of creating many vectors
to study the interactions of genes, we aimed to create a plasmid with a designed circuit that
would control the transcription of several tuberculosis genes using invertases. We chose to
use non-pathogenic mycobacterium tuberculosis genes and transcription factors within non-
pathogenic E.Coli for safety reasons.
The genetic mechanisms that trigger the change between the two
forms are not very well understood. The genetic network of tuberculosis is complicated and
studying the interactions between genes can be time consuming, requiring the construction of
many plasmids to study only one small part of the network. To look for new, faster ways of
studying this problem we turned to synthetic biology. Instead of creating many vectors
to study the interactions of genes, we aimed to create a plasmid with a designed circuit that
would control the transcription of several tuberculosis genes using invertases. We chose to
use non-pathogenic mycobacterium tuberculosis genes and transcription factors within non-
pathogenic E.Coli for safety reasons.
Invertases are recombinases that will bind to recognition sites. Once bound, the DNA between two sites will invert and flip horizontally. This effectively acts as an on/off switch for transcription of that region of DNA as it is not available for DNA polymerase to bind to. In terms of circuit design, it will act as a not gate. Using them within a plasmid with several tuberculosis genes would allow us to check for regulatory measures such as negative feedback, etc.

An example of a construct built around the invertase architecture.
In addition to the novel cellular architecture, we also used a variety of software tools created by our computational teams (Boston University, Wellesley):
- G-nome Surfer Pro
G-nome Surfer Pro is an integrated environment that allows for the viewing of prokaryotic genomic data and literature associated with the genome. As it is built on a Microsoft Surface, it encourages collaboration. We used the Primer Genie to design the primers for the tuberculosis genes and transcription factors. - Trumpet
Trumpet consists of a library of genes and promoters, which can be configured into any desired permutation or combination by treating the DNA with recombinases, which allows us to rewire the genes and switches and study all their combinations. It was used by the wetlab to help us map out where the invertases should go in our plasmids in order to correctly turn on and off the segments of DNA we want to study. - PuppetShow
This suite includes a high level programming language for specifying biological protocols commonly used in the laboratory, which are then executed by a liquid-handling robot with minimal user intervention. We used this in conjunction with the robot (described below) to create samples that were simultaneously created manually to compare the results. - Clotho
Clotho is used to mange, create and store new biological building blocks in community based repositories. It includes a suite of tools that include PuppetShow and Trumpet, which were designed specifically for this project. Other tools that were used were:- Bull Trowel
- Spreadit Parts
- Spreadit Vectors
- Spreadit Features
- Optimus Primer-primer designer
- Feature Chomp-reads in APE and GENBANK files and takes the features found within the file to a feature database
- Batterboard-allowed us to electronically represent physical samples in the lab
Another way we looked into facilitating progress in studying genetic networks is through the use of automation:
- Robot The liquid handling robot was used to reproduce molecular biology protocols in a more efficient and accurate manner. The robot is controlled by Puppeteer which creates code necessary to choose the correct series of commands in EVOware,the robot's software.
- QIA Cube
Results
Being associated with a large computational team, the wetlab team was unique as our results not only consisted of creating novel forms of DNA, but evaluating the usefulness of the software tools our computational team produced. Our success was not only measured in the number of colonies we could get to grow, but also in how well we could incorporate these tools and give informative feedback.
The first vectors we were interested in creating were fluorescent protein reporters. Our goal was to eventually fuse genes with the reporter devices in order to establish that the genes were being turned on or off, but we first wanted to characterize the luminescence of the reporting devices. We used Clotho to store the pieces of DNA we were hoping to create, listed here:
Parts and diagrams.
This list of parts would give us red, green, yellow, and blue colors to play with. We then set out to create the devices.
Approach
Lorem ipsum dolor sit amet, consectetur adipiscing elit. Quisque ut tincidunt quam. Praesent in sodales mi. In hac habitasse platea dictumst. Nunc magna massa, gravida in euismod nec, rutrum sed dolor. Sed accumsan ullamcorper quam nec feugiat. Nam interdum volutpat nibh nec ultrices. Pellentesque habitant morbi tristique senectus et netus et malesuada fames ac turpis egestas. Donec eget nibh leo. Nullam euismod vulputate leo, eu egestas dolor volutpat at. Vivamus ac purus mi, vitae consequat enim.
 "
"
