Team:Paris Bettencourt/Modeling/tRNA diffusion
From 2011.igem.org
| Line 46: | Line 46: | ||
| - | <h4>Receiver gene construct | + | <h4>Receiver gene construct - T7 amber</h4> |
| - | <p>The receiver | + | <p>The receiver gene construct is modeled by the following equations:</p> |
| - | <img src='https://static.igem.org/mediawiki/2011/ | + | <img src='https://static.igem.org/mediawiki/2011/e/ee/TRNA_T7_amber.png' style='width:100%;'> |
<br> | <br> | ||
| + | <p>You will remark that activated <i>mRNA<sub>amber</sub></i> is noted <i>mRNA<sub>amber</sub><sup>*</sup></i>.</p> | ||
<br> | <br> | ||
<p>The reporter for the receiver and amplification gene construct (<i>GFP</i>) is modeled by the following equations:</p> | <p>The reporter for the receiver and amplification gene construct (<i>GFP</i>) is modeled by the following equations:</p> | ||
Revision as of 15:34, 8 September 2011

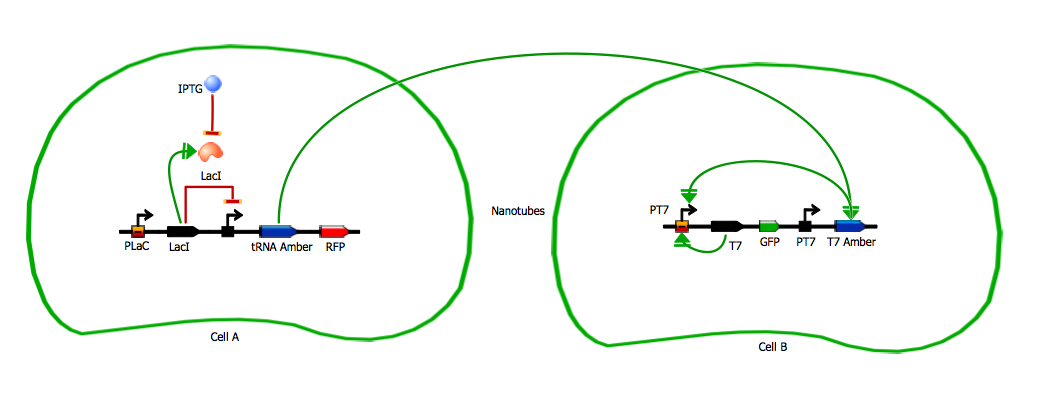
Model for tRNA amber diffusion system
Summary
The tRNA amber diffusion design is the design we have using the smallest molecule we want to diffuse through nanotubes: tRNA. The emitter cell produces this tRNA amber which is transfered through nanotubes and serves to translate the so-called mRNA amber (mRNA for T7 RNA polymerase with two amber mutations). This mRNA amber can be translated only in presence of our mutant tRNA and is therefore our recetor construct. Having two amber mutations in the T7 RNA polymerase gene in the receiver cell should prevent any leakiness from the receiver construct. The auto-amplification is obtained through the same T7 auto-amplification loop used in the T7 RNA polymerase diffusion design.
The behaviour expected is to have an ON/OFF switch activating when a certain number of tRNA amber have entered the cell. To be certain this activation was possible, we ran additionnal simulation of tRNA diffusion in one cell. Our conclusion was that a delay of a less than 10 minutes might be observed in the receiver cell if very few tRNA amber arrive in the receiver cell. You can find more about this sub-model here.
You will find below the results for a simple simulation with every part of the design in one cell. All of our components are in one cell. Between t=7500s and t=12500s, IPTG is added to the cell, lifting repression by LacI. RFP is the reporter for the emitting construct and GFP for the receiving construct.

Our simulation exhibits the behaviour we expected. The model shows that in our experimental conditions the cell should produce a significant response in a reasonable time (approximately 20 to 30 minutes after the tRNA amber appears in the cell). Once started, the auto-amplification loop can not be stopped, as we can see with the GFP staying at its maximum even when RFP levels decrease. The major limit of this model could come from leakiness of promoters. If the pT7 is not as reliable as we thought, the auto-amplification loop could trigger itself all the time.
Even though the timescale for tRNA amber diffusion in the receiver cell is reasonable, we tried several possible stable states of tRNA amber input for the receiver construct. This stable state hypothesis for tRNA amber is justified by the recycling of tRNA in the cells.
Design
The tRNA amber diffusion design is the design we have using the smallest molecule we want to diffuse through nanotubes: tRNA. The emitter construct produces tRNA amber a tRNA which is complementary to amber stop codons but transports an amino-acid (TO BE COMPLETED) instead of stopping translation. tRNA amber is then transfered through the nanotubes and into the receiver cell.
In the receiver cell, a T7 RNA polymerase amber gene constitutively produces the so-called mRNA amber (mRNA for T7 RNA polymerase with two amber mutations) which is our receiver construct. This mRNA amber can be translated into functional T7 RNA polymerase only in presence of our mutant tRNA amber. We note the functional polymerases expressed from this gene T7 amber.
The amplifier construct is then a T7 auto-amplifying loop identical to the one in the T7 RNA polymerase diffusion design. The T7 amber activates this loop which is also where our reporter GFP is expressed.
This construction was put in two different settings. One is what we just described, where the emitting gene network is in one cell and the receiving gene network is in another. In the other construction, everything is in one cell. We use the second construct as a control to really see the impact of the cell-to-cell communication on the behaviour of the cells.
We ran our models for those two configurations. We used a steady flow of signaling molecules in the receiver cell for the "one emitting cell - one receiving cell" construction. You can find our justifications about this assumption here.
Model
LacI
We use LacI as a repressor for the emitter gene construct. LacI repression can be cancelled by IPTG. This way we can induce production of RFP and tRNA amber by adding IPTG on the cells.


Inactivated LacI can not repress the pLAC promoter anymore. Note that we consider that the reaction between IPTG and LacI fires without any delay. This assumption is justified by the fact that this reaction is much faster than any other in our gene network.
Emitter gene construct - tRNA amber
The emitter gene construct is modeled by the following equations:

The reporter for the emitter gene construct (RFP) is modeled by the following equations:

Receiver gene construct - T7 amber
The receiver gene construct is modeled by the following equations:

You will remark that activated mRNAamber is noted mRNAamber*.
The reporter for the receiver and amplification gene construct (GFP) is modeled by the following equations:

Parameters
This design relies on tRNA amber as the signaling molecule going through the nanotubes.
The parameters used in this model are:
| Parameter | Description | Value | Unit | Reference |
|---|---|---|---|---|
 |
Active LacI concentration (LacI which is not inactivated by IPTG) | NA | molecules per cell |
Notation convention |
 |
IPTG concentration | NA | molecules per cell |
Notation convention |
 |
Inactived LacI concentration | NA | molecules per cell |
Notation convention |
 |
Total LacI concentration | TBD | molecules per cell |
Steady state for equation |
 |
T7 RNA polymerase (emitter, T7') concentration | NA | molecules per cell |
Notation convention |
 |
mRNA associated with T7' concentration | NA | molecules per cell |
Notation convention |
 |
T7 RNA polymerase (auto-amplification, T7'') concentration | NA | molecules per cell |
Notation convention |
 |
mRNA associated with T7'' concentration | NA | molecules per cell |
Notation convention |
 |
Maximal production rate of pVeg promoter (constitutive) | ??? | molecules.s-1 or pops |
Estimated |
 |
Maximal production rate of pLac promoter | 0.02 | molecules.s-1 or pops |
Estimated |
 |
Maximal production rate of pT7 promoter | 0.02 | molecules.s-1 or pops |
Estimated |
 |
Dissociation constant for IPTG to LacI | 1200 | molecules per cell |
Aberdeen 2009 wiki |
 |
Dissociation constant for LacI to LacO (pLac) | 700 | molecules per cell |
Aberdeen 2009 wiki |
 |
Dissociation constant for T7 RNA polymerase to pT7 | 3 | molecules per cell |
Estimated ADD EXPLANATION |
 |
Translation rate of proteins | 1 | s-1 | Estimated ADD EXPLANATION |
 |
Dilution rate in exponential phase | 2.88x10-4 | s-1 | Calculated with a 40 min generation time. See explanation |
 |
Degradation rate of mRNA | 2.88x10-3 | s-1 | Uri Alon (To Be Confirmed) |
 |
Delay due tT7 RNA polymerase production and maturation | 300 | s | http://mol-biol4masters.masters.grkraj.org/html/Prokaryotic_DNA_Replication13-T7_Phage_DNA_Replication.htm |
 |
Delay due to mRNA production | 30 | s | http://bionumbers.hms.harvard.edu/bionumber.aspx?s=y&id=104902&ver=5&hlid=58815 2kb/(50b/s) --> approximation: all our contructs are around 2kb |
Results & discussions
Limits
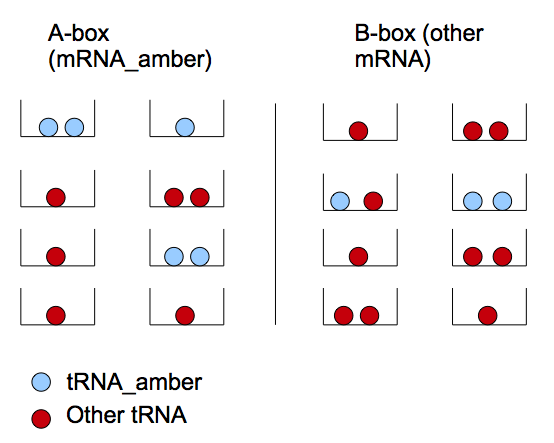
The amber suppressor tRNA diffusion. The idea of the system is to pass tRNA amber molecules through the nanotubes. At every moment of time in the receiver cell there is a certain amount of transcribed mRNA-T7 among the others mRNA. The behavior of tRNA amber that arrived in a receiver cell is random, so in order to describe its interaction with mRNA-T7 and its further translation we can reason in terms of probability.
We can reason in two steps : first a tRNA amber molecule gets close to a mRNA molecule. Then, it binds it's anti-codon with a codon of the mRNA. This reasoning is similar to the problem of boxes and balls. There are two types of boxes: 'a' of the first type and 'b' of the second (which corresponds to the set of mRNA-T7 and mRNA-non-T7), and there are 't' balls(tRNA amber). All the balls are randomly distributed in the boxes. If there are two or more balls in some box of the first type (two or more tRNA amber per mRNA-T7) then a T7 molecule will be produced with a chance P_0.
We have defined two models for this system which both rely on the following assumptions :
- Each mRNA is defined as a 'box'
- All the tRNA molecules are uniformly distributed in the boxes.
- The number of tRNA_amber diffused through the nanotubes is much more smaller than the one of the mRNA. Thus the chance that three or more tRNA amber will "find" one mRNA-T7 is negligible comparing to the one of two tRNA amber (finding a mRNA-T7). In our model we will consider that at one moment of time each mRNA interacts with 0, 1 or 2 tRNA ambers.
- The tRNA_amber placed in a correct box are always used
We have defined two models for this system which both rely on the following assumptions :
- Each mRNA is defined as a 'box'
- All the tRNA molecules are uniformly distributed in the boxes.
- The number of tRNA_amber diffused through the nanotubes is much more smaller than the one of the mRNA. Thus the chance that three or more tRNA amber will "find" one mRNA-T7 is negligible comparing to the one of two tRNA amber (finding a mRNA-T7). In our model we will consider that at one moment of time each mRNA interacts with 0, 1 or 2 tRNA ambers.
- The tRNA_amber placed in a correct box are always used
Let's define two types of boxes: mRNA_amber ('A') and other type of mRNA ('B'). We note the mRNA_amber producing T7 as mRNA*_amber. The latter appears if we have two tRNA_amber in one box. This model treats the repartition of tRNA_amber in the different boxes.
We note P(x) the probability of having x A-boxes containing two tRNA_amber. Thus P(x=1) corresponds to the probability of finding a couple of tRNA_amber in an A-box, thus to produce x T7 molecules. 't' is the number of tRNA_amber in the cell.
We note:
- P(x=1)= (probability that 2 balls choose A-boxes) * (probability that these 2 balls choose the same A-box) + (probability that 3 balls choose A boxes) * (probability that 2 balls out of 3 choose the same A-box and the third doesn't) + ... + (probability that t balls choose an A-box) * (probability that 2 of these t balls choose the same A-box and the rest don't).
- P(x=2)= (probability that 4 balls choose A-boxes) * (probability that these 4 balls choose 2 A-boxes, one A-box per pair of balls) + (probability that 5 balls choose A-boxes) * (probability that 4 balls out of 5 choose 2 A-boxes, one A-box per pair of balls and the fifth doesn't) + ... + (probability that t balls choose A-boxes) * (probability that 4 out of these t balls choose [t/2] A-boxes, one A-box per pair of balls and the rest don't).
- ...
- P(x=i)= (probability that 2i balls choose A-boxes) * (probability that these 2i balls choose i A-boxes, one A-box per pair of balls) + (probability that (2i + 1) balls choose A-boxes) * (probability that 2i balls out of (2i + 1) choose i A-boxes, one A-box per pair of balls and the rest don't) + ... + (probability that t balls choose A-boxes) * (probability that 2i out of these t balls choose [t/2] A-boxes, one A-box per pair of balls and the rest don't).
Hence:
 "
"