Team:British Columbia/Modeling
From 2011.igem.org
(→MODEL 1: Secretion and Production of Monoterpenes in Yeast) |
(→MODEL 1: Secretion and Production of Monoterpenes in Yeast) |
||
| Line 6: | Line 6: | ||
<p></p> | <p></p> | ||
| - | This modeling component of our project was to simulate the biochemical pathways for our genetically engineered yeasts. Simulations were programmed using MATLAB. Every yeast cell is designed to produce one specific monoterpene in mass, and others monoterpenes in small amounts. Each simulation is based on differential equations that were made based on using first order and Michelson-menten kinetics. Our project is based on research that has been conducted in 2011 so some variables will need to be experimentally determined in the future to make more accurate simulations | + | This modeling component of our project was to simulate the biochemical pathways for our genetically engineered yeasts. Simulations were programmed using MATLAB. Every yeast cell is designed to produce one specific monoterpene in mass, and others monoterpenes in small amounts. Each simulation is based on differential equations that were made based on using first order and Michelson-menten kinetics. Our project is based on research that has been conducted in 2011 so some variables will need to be experimentally determined in the future to make more accurate simulations. |
Revision as of 18:44, 5 September 2011

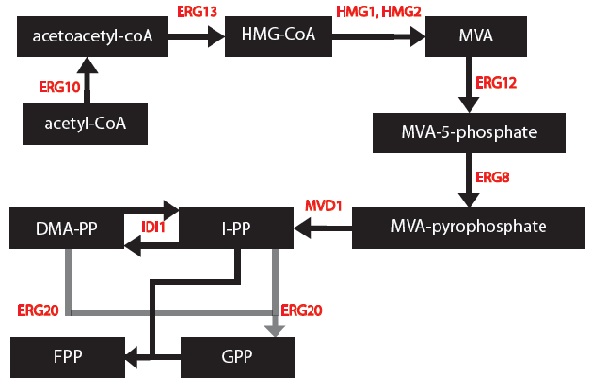
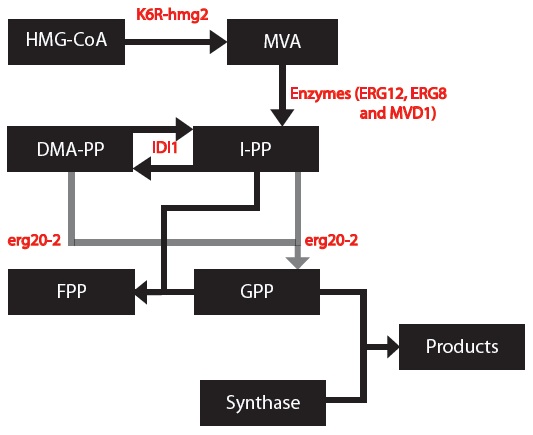
MODEL 1: Secretion and Production of Monoterpenes in Yeast
OBJECTIVE: To develop a model to assay the secretion and production of monoterpenes in yeast.
This modeling component of our project was to simulate the biochemical pathways for our genetically engineered yeasts. Simulations were programmed using MATLAB. Every yeast cell is designed to produce one specific monoterpene in mass, and others monoterpenes in small amounts. Each simulation is based on differential equations that were made based on using first order and Michelson-menten kinetics. Our project is based on research that has been conducted in 2011 so some variables will need to be experimentally determined in the future to make more accurate simulations.
We made a list of important common chemical reactions after developing this simplified version of the biochemical pathway:
Since each synthase gene combines with GPP to produce different monoterpenes, some chemical reactions will be specific to a certain synthase.
The chemical equations specific to the PgTPS-Pin2 synthase gene:
The chemical equations specific to the PsTPS-Car1 synthase gene:
The chemical equations specific to the PgxeTPS-Cin synthase gene:
The chemical equations specific to the PgTPS-Cin synthase gene:
The chemical equations specific to the PsTPS-Pin synthase gene:
Next, we created differential equations from the series of chemical reactions and block diagrams for each of the biochemical pathways.
These are the differential equations that are common to each biochemical pathway:
Since each synthase gene combines with GPP to produce different monoterpenes, some differential equations will be specific to a certain synthase.
The differential equations specific to the PgTPS-Pin2 synthase gene:
The differential equations specific to the PsTPS-Car1 synthase gene:
The differential equations specific to the PgxeTPS-Cin synthase gene:
The differential equations specific to the PgTPS-Cin synthase gene:
The differential equations specific to the PsTPS-Pin synthase gene:
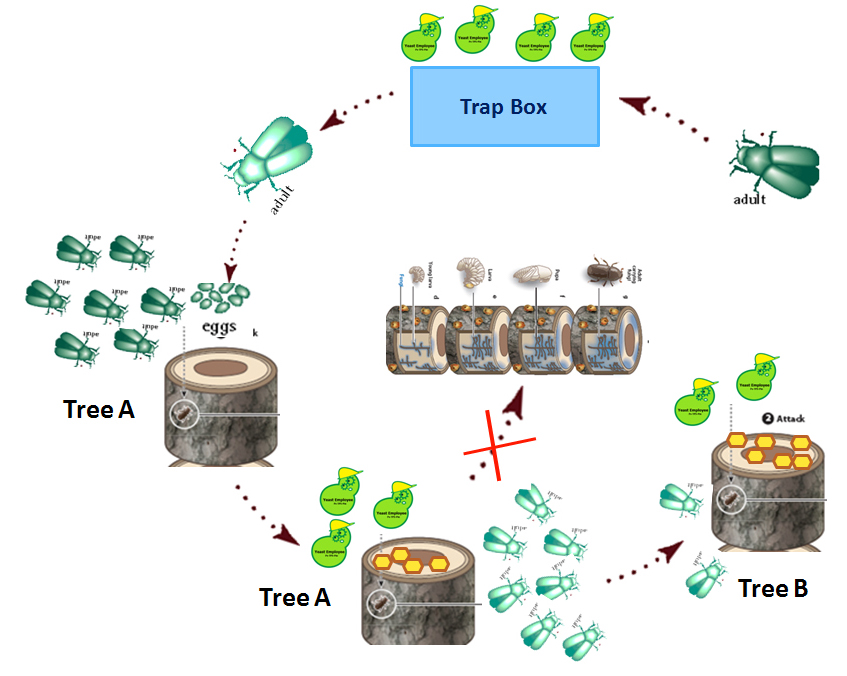
MODEL 2: Dynamics of Mountain Pine Beetle Populations in British Columbia
OBJECTIVE: To construct a mathematical model that can predict the dynamics of the mountain pine beetle populations in B.C. under the influence of our synthetic product. This model can be coupled with the strategic planning for future conservation initiatives.
We will show how the pine beetle is predicted to spread in British Columbia, according to the government of British Columbia, and compare that to how we predict our modified yeasts may slow down the spread of the pine beetle.
 "
"