Team:Tsinghua/project
From 2011.igem.org
(→Project Overview) |
(→Movement) |
||
| (61 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | { | + | __NOTOC__ |
| - | + | {{:Team:Tsinghua/header}}<html> | |
| - | + | <body id="bd_home"> | |
| - | + | </html> | |
| - | + | {{:Team:Tsinghua/leftbar}} | |
| - | + | <html> | |
| - | + | <div id="content_box"> | |
| - | + | <script> | |
| - | + | navl = new Array("Overview", "Experiments", "Modeling"); | |
| + | lk = new Array("#overview", "/Team:Tsinghua/experiment", "/Team:Tsinghua/modeling"); | ||
| + | writenav(navl, lk); | ||
| + | </script> | ||
| - | = | + | <div id="main_content"> |
| - | + | </html> | |
| - | + | <html><a id="overview"></a></html> | |
| - | + | =E. Colimousine= | |
| + | ==Overview== | ||
| - | + | Single cells can be powerful in information processing, but information processors assembled from just a few cells are limited in their capabilities. In a complex information processing system, separation of sensor and processor is necessary for eliminating unwanted cross-talk, which in turn requires the signal transduction system. | |
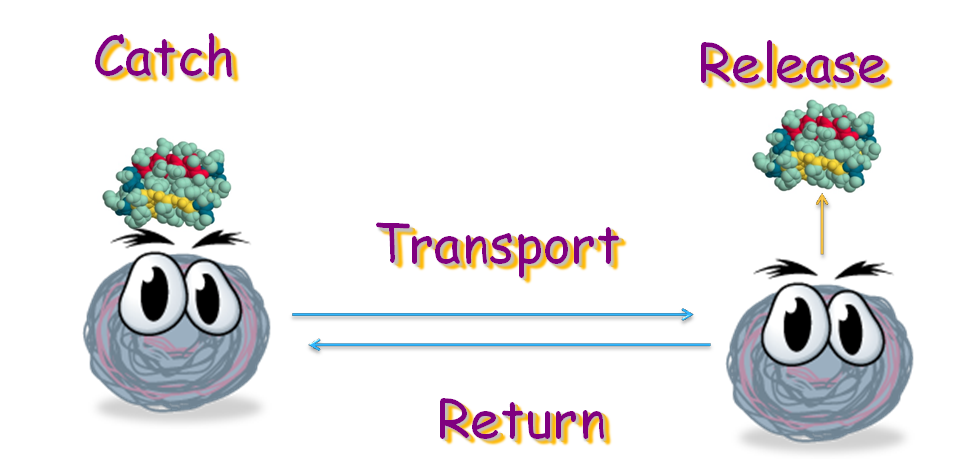
| - | + | Our project is destined to generate an ''E. coli'' strain which can transport signal protein along a gradient back and forth. The bacteria can bind with the target protein at one place and then, by sensing the gradient of certain amino acid, move to its destination, where it releases the protein with the help from another ''E. coli'' strain. When this is done, it will move back to transport more target protein. In this process, four modules are necessary: binding, releasing, and movement. | |
| - | + | [[File:Sketch.png|500px]] | |
| - | + | ==Binding== | |
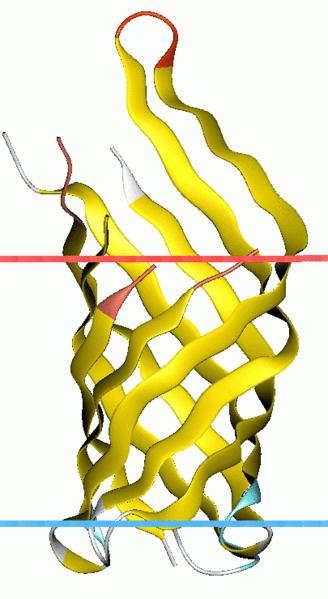
| - | + | We first need to conjugate a binding vehicle directly sensing substrate in the medium. <html><span id="a-tip">Outer membrane protein A (OmpA) <a class="div-tip">OmpA-like transmembrane domain is an evolutionarily conserved domain of outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated by a variety of mechanisms.</a></span></html> comes into our notice for its outstanding property that proteins linked to its C-terminal will be pushed out of the bacteria’s membrane. | |
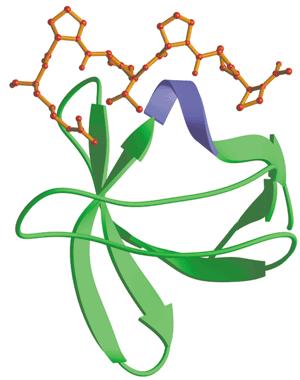
| + | <html><span id="a-tip">Src-homology 3(SH3) domain<a class="div-tip">Src-homology 3 (SH3) domain has high affinity for proline-rich peptides and together they can form a left-handed poly-Pro type II helix, with the minimal consensus Pro-X-X-Pro.</a></span> </html> which has high affinity for proline-rich peptides, could act like tongs to grab any protein with a proline riich motif. For easy track, fluorescent protein mCherry was the one linked withthe binding motif (Kd=3.67μM). | ||
| - | + | <gallery heights=250px widths=250px perrow=2> | |
| + | File:SH3_domain.jpg|SH3 binding domain (shown bound to proline-rich peptide) | ||
| + | File:OmpA-like_transmembrane_domain.jpg|OmpA transmembrane segment | ||
| + | </gallery> | ||
| - | + | The binding module eventually comprises of two proteins, namely, OmpA-SH3 protein, which functions as the binding vehicle and proline-rich containing mCherry protein, which functions as the binding substrate. | |
| - | + | ||
| - | + | ||
| + | ==Releasing== | ||
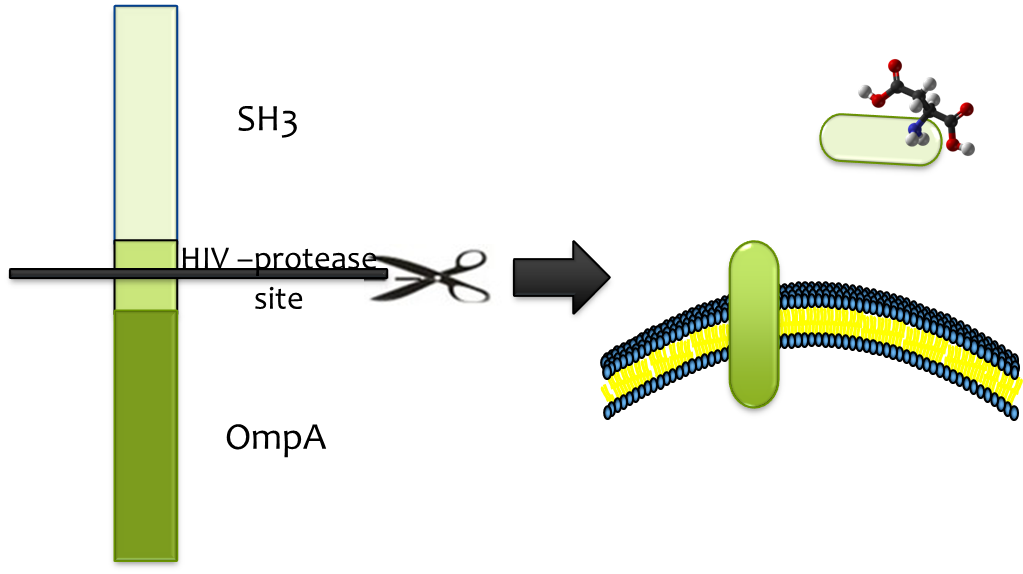
| - | + | Considering of the significance of efficient and specific release from strong binding, HIV-protease site was constructed into the binding cassette, which enables proteolytic release by <html><span id="a-tip">HIV-protease<a class="div-tip"> HIV-protease is readily available and its high efficiency, low molecular weight and high specificity matches the fine candidate</a> </span></html>. We fused HIV-protease with OmpA and expressed it in a second ''E. coli'' strain which we immobilize at the destination, so substrate (together with SH3 domain) could be released once the E. colimousine arrives there. | |
| - | + | <gallery widths=300px heights=200px> | |
| + | File:SH3-HIV_protease_site-OmpA.png|Protease site in binding cassete | ||
| + | File:SH3-HIV_protease_site-OmpA_final.png|Proteolytic release | ||
| + | </gallery> | ||
| - | + | ==Movement== | |
| - | + | Regulatory network controlling ''E. coli'' chemotaxis is one of the well-established models of signaling transduction, hence chemotaxis is most frequently used in guiding the movement of ''E. coli'' cells. Here, receptors Tar and Tsr* are assembled into large clusters that are predominantly located at the poles of the bacterium could sense extracellular signals as aspartate and serine respectively. The CheW protein binds to the receptors and acts as a scaffold protein to recruit CheA, whose phosphorylation is stimulated by the receptor. The phosphate is transferred to the cytoplasmic protein CheY, which controls flagellar rotation and in turn, controls the movement of E.coli. Previously, a pair of mutations in CheW* (V108M) and Tsr* (E402A) was shown to produce an orthogonal pair (R.B. Bourret, A.M. Stock, ''et al''., 2002), where the mutated proteins interact with each other but not with wildtype Tsr or CheW. So, this can be used to toggle between activating the serine-responsive Tsr* and aspartate-responsive Tar. When we set up two gradients (serine and aspartate) in the opposite direction, the bacteria can move forth and back. | |
| + | |||
| + | [[File:Thudgrad1.png|300px]] | ||
| + | [[File:Thudgrad2.png|300px]] | ||
| + | |||
| + | As leakage level expression of chemotaxis receptor is sufficient to drive chemotaxis, we resorted to pChemoK plasmid constructed in Christopher's lab (T.S. Moon, E.J. Clarke, ''et al''., 2011). In this plasmid, expression of Tar and Tsr is controled by fimE invertase system. As the direction of promoter is inverted, no basal transcription exists and strict control of expression is achieved. | ||
| + | |||
| + | [[File:Thuchemok.png|200px|pChemoK]] | ||
| + | |||
| + | ''pChemoK plasmid, from Christopher's lab'' | ||
| + | |||
| + | |||
| + | |||
| + | <html></div></div> | ||
| + | |||
| + | |||
| + | <html></div></div> | ||
Latest revision as of 17:36, 28 October 2011

E. Colimousine
Overview
Single cells can be powerful in information processing, but information processors assembled from just a few cells are limited in their capabilities. In a complex information processing system, separation of sensor and processor is necessary for eliminating unwanted cross-talk, which in turn requires the signal transduction system.
Our project is destined to generate an E. coli strain which can transport signal protein along a gradient back and forth. The bacteria can bind with the target protein at one place and then, by sensing the gradient of certain amino acid, move to its destination, where it releases the protein with the help from another E. coli strain. When this is done, it will move back to transport more target protein. In this process, four modules are necessary: binding, releasing, and movement.
Binding
We first need to conjugate a binding vehicle directly sensing substrate in the medium. Outer membrane protein A (OmpA) OmpA-like transmembrane domain is an evolutionarily conserved domain of outer membrane proteins. This domain consists of an eight-stranded beta barrel. OmpA is the predominant cell surface antigen in enterobacteria found in about 100,000 copies per cell. The expression of OmpA is tightly regulated by a variety of mechanisms. comes into our notice for its outstanding property that proteins linked to its C-terminal will be pushed out of the bacteria’s membrane.
Src-homology 3(SH3) domainSrc-homology 3 (SH3) domain has high affinity for proline-rich peptides and together they can form a left-handed poly-Pro type II helix, with the minimal consensus Pro-X-X-Pro. which has high affinity for proline-rich peptides, could act like tongs to grab any protein with a proline riich motif. For easy track, fluorescent protein mCherry was the one linked withthe binding motif (Kd=3.67μM).
The binding module eventually comprises of two proteins, namely, OmpA-SH3 protein, which functions as the binding vehicle and proline-rich containing mCherry protein, which functions as the binding substrate.
Releasing
Considering of the significance of efficient and specific release from strong binding, HIV-protease site was constructed into the binding cassette, which enables proteolytic release by HIV-protease HIV-protease is readily available and its high efficiency, low molecular weight and high specificity matches the fine candidate . We fused HIV-protease with OmpA and expressed it in a second E. coli strain which we immobilize at the destination, so substrate (together with SH3 domain) could be released once the E. colimousine arrives there.
Movement
Regulatory network controlling E. coli chemotaxis is one of the well-established models of signaling transduction, hence chemotaxis is most frequently used in guiding the movement of E. coli cells. Here, receptors Tar and Tsr* are assembled into large clusters that are predominantly located at the poles of the bacterium could sense extracellular signals as aspartate and serine respectively. The CheW protein binds to the receptors and acts as a scaffold protein to recruit CheA, whose phosphorylation is stimulated by the receptor. The phosphate is transferred to the cytoplasmic protein CheY, which controls flagellar rotation and in turn, controls the movement of E.coli. Previously, a pair of mutations in CheW* (V108M) and Tsr* (E402A) was shown to produce an orthogonal pair (R.B. Bourret, A.M. Stock, et al., 2002), where the mutated proteins interact with each other but not with wildtype Tsr or CheW. So, this can be used to toggle between activating the serine-responsive Tsr* and aspartate-responsive Tar. When we set up two gradients (serine and aspartate) in the opposite direction, the bacteria can move forth and back.
As leakage level expression of chemotaxis receptor is sufficient to drive chemotaxis, we resorted to pChemoK plasmid constructed in Christopher's lab (T.S. Moon, E.J. Clarke, et al., 2011). In this plasmid, expression of Tar and Tsr is controled by fimE invertase system. As the direction of promoter is inverted, no basal transcription exists and strict control of expression is achieved.
pChemoK plasmid, from Christopher's lab
 "
"