Team:NTNU Trondheim/LacI+pLac+mCherry
From 2011.igem.org
m (→Stress sensor precursor (LacI+pLac+mCherry)) |
m (→Characterization) |
||
| (15 intermediate revisions not shown) | |||
| Line 10: | Line 10: | ||
Our experiments indicate that the stress sensor precursor is put together the right way. | Our experiments indicate that the stress sensor precursor is put together the right way. | ||
The stress sensor precursor is made from the following biobricks: | The stress sensor precursor is made from the following biobricks: | ||
| + | |||
| + | [[File: Super_BB__minus_p1_1-5-2.jpg|thumb|Five parallels digested with BsaAI + HpaI and BstBI SpeI]] | ||
| + | |||
{|border="1" | {|border="1" | ||
!Biobrick | !Biobrick | ||
| Line 27: | Line 30: | ||
|} | |} | ||
| - | The composite biobrick has a total length of 2153 bp, and | + | |
| + | The composite biobrick has a total length of 2153 bp, and was originally located in the plasmid pSB1A2. In order the check if the construct was correct we digested the plasmid with BsaAI + HpaI and BstBI SpeI. Prior to shipping the biobrick was transfered to the official shipping plasmid pSB1C3. | ||
This construct could in theory be used for any kind of biosensor. The only thing needed is a promotor that gets downregulated directly or indirectly by the conditions one would want to detect. | This construct could in theory be used for any kind of biosensor. The only thing needed is a promotor that gets downregulated directly or indirectly by the conditions one would want to detect. | ||
| + | |||
| + | |||
| + | ==Characterization== | ||
| + | |||
| + | The part gives red culture and red colonies when grown in LB and M9, and on LA. | ||
| + | |||
| + | '''Sequencing''' | ||
| + | |||
| + | |||
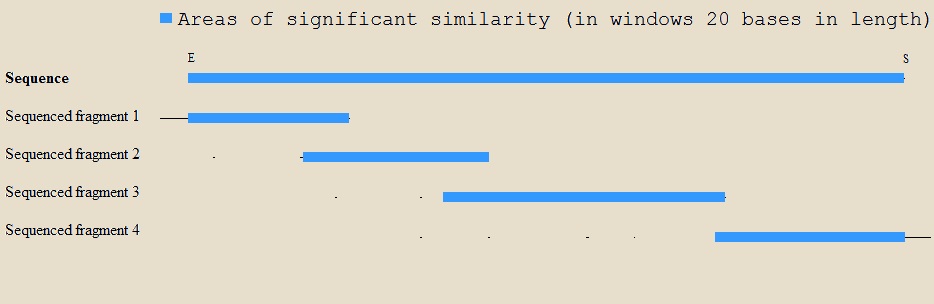
| + | The BioBrick's sequence was confirmed by sequencing of the plasmid containing it. A map of the alignment of the sequencing fragments to the BioBrick's sequence is shown below. All bases were checked in detail. Prefix and suffix sequences were also correct, but are not shown in the map. | ||
| + | |||
| + | |||
| + | [[File:Brick-P.jpg|747px]] | ||
| + | |||
| + | |||
| + | '''Link to parts registry:''' [http://partsregistry.org/Part:BBa_K639000 BBa_K639000] | ||
{{:Team:NTNU_Trondheim/NTNU_footer}} | {{:Team:NTNU_Trondheim/NTNU_footer}} | ||
Latest revision as of 15:30, 21 September 2011

Stress sensor precursor (LacI+pLac+mCherry)
This is our system without the rrnB P1-promotor. Because this construct lacks a downregulated promotor in front of it, LacI will not be syntethized, and in response, pLac will not be downregulated of LacI. Since pLac is not downregulated, mCherry is going to be present whether the cells are stressed or not. Our experiments indicate that the stress sensor precursor is put together the right way. The stress sensor precursor is made from the following biobricks:
| Biobrick | Part number |
|---|---|
| LacI with RBS | BBa_J24679 |
| Terminator | BBa_B0015 |
| pLac | BBa_R0011 |
| mCherry with RBS and terminator | BBa_J06702 |
The composite biobrick has a total length of 2153 bp, and was originally located in the plasmid pSB1A2. In order the check if the construct was correct we digested the plasmid with BsaAI + HpaI and BstBI SpeI. Prior to shipping the biobrick was transfered to the official shipping plasmid pSB1C3.
This construct could in theory be used for any kind of biosensor. The only thing needed is a promotor that gets downregulated directly or indirectly by the conditions one would want to detect.
Characterization
The part gives red culture and red colonies when grown in LB and M9, and on LA.
Sequencing
The BioBrick's sequence was confirmed by sequencing of the plasmid containing it. A map of the alignment of the sequencing fragments to the BioBrick's sequence is shown below. All bases were checked in detail. Prefix and suffix sequences were also correct, but are not shown in the map.
Link to parts registry: [http://partsregistry.org/Part:BBa_K639000 BBa_K639000]
 "
"