Team:Edinburgh/Cellulases (C model)
From 2011.igem.org
| (110 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Edinburgh/tech/Navbox}} | {{:Team:Edinburgh/tech/Navbox}} | ||
<html><script type="text/javascript">$(document).ready(function() { | <html><script type="text/javascript">$(document).ready(function() { | ||
| - | getMenus('model', ' | + | getMenus('model', 'model_synergyc'); |
}); </script></html> | }); </script></html> | ||
<div class="main_body"> | <div class="main_body"> | ||
| - | |||
| - | |||
| - | # Exoglucanase chews away at the end of a | + | <p class="h1">Cellulases (C model)</p> |
| + | |||
| + | As explained in the abstract, the feasibility of our improved biorefineries depends on exploiting <span class="hardword" id="synergy">synergy</span> between enzymes. Therefore, we attempted to use the C programming language to construct a model which could prove that synergy is, or might be, a real phenomenon affecting <span class="hardword" id="cellulase">cellulase</span> action. | ||
| + | |||
| + | ==Synergy explained== | ||
| + | |||
| + | It can seem almost magical that having enzymes closer together can increase their usefulness, but consider the following statements about <span class="hardword" id="cellulose">cellulose</span> degradation, all of which are true ([http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-091208-085603 Fontes and Gilbert, 2010]): | ||
| + | |||
| + | # Exoglucanase chews away at the end of a cellulose chain, producing <span class="hardword" id="cellobiose">cellobiose</span> sugars. | ||
# Endoglucanase cuts cellulose chains in the middle, essentially turning one chain into two. | # Endoglucanase cuts cellulose chains in the middle, essentially turning one chain into two. | ||
# Cellobiose inhibits the action of the above enzymes. | # Cellobiose inhibits the action of the above enzymes. | ||
| Line 16: | Line 22: | ||
==''Syn:'' a simple demonstration of synergy== | ==''Syn:'' a simple demonstration of synergy== | ||
| - | We can demonstrate that there is (potentially) a huge difference between the synergistic and non-synergistic systems with Edinburgh's ''Syn'' | + | We can demonstrate that there is (potentially) a huge difference between the synergistic and non-synergistic systems with Edinburgh's ''Syn'' program, written in C. This is a 2D simulation that works in the following way: |
* The world is a 2D grid. | * The world is a 2D grid. | ||
** Each spot in the grid can contain a '''sugar''', a '''bond''' between sugars, or '''nothing'''. | ** Each spot in the grid can contain a '''sugar''', a '''bond''' between sugars, or '''nothing'''. | ||
** The world begins with an array of cellulose. | ** The world begins with an array of cellulose. | ||
| - | * Cellulose is modelled as alternating sugars and bonds: ''s-b-s-b-s-b-s-b-s-b-s'' | + | * Cellulose is modelled as horizontal chains with alternating sugars and bonds: ''s-b-s-b-s-b-s-b-s-b-s'', etc. |
* There are three enzyme types: '''endoglucanase''', '''exoglucanase''', and '''β-glucosidase'''. | * There are three enzyme types: '''endoglucanase''', '''exoglucanase''', and '''β-glucosidase'''. | ||
| - | ** The enzymes move about randomly | + | ** The enzymes move about randomly in manner similar to <span class="hardword" id="brownian">Brownian motion</span>. |
| - | ** If an enzyme is in the same place as a bond, it can | + | ** If an enzyme is in the same place as a bond, it can destroy it: |
| - | *** Endoglucanase | + | *** Endoglucanase only cuts bonds away from the ends of a chain (some minimum distance is specified). |
| - | *** Exoglucanase | + | *** Exoglucanase only cuts bonds if this results in a cellobiose molecule (''s-b-s'') forming. |
| - | *** β-glucosidase | + | *** β-glucosidase only cuts cellobiose bonds. |
* There is inhibition of exoglucanase by cellobiose; it does not cut bonds (or has a reduced chance to do so) if there is a nearby cellobiose molecule. | * There is inhibition of exoglucanase by cellobiose; it does not cut bonds (or has a reduced chance to do so) if there is a nearby cellobiose molecule. | ||
| + | So, the actions of the enzymes are as follows: | ||
| - | [[Image:Edinburgh-All-Actions.png|thumb|center|620px|Sugars are dark green squares, bonds between sugars are light green. From left to right are shown the actions of endoglucanase, exoglucanase, and β-glucosidase.]] | + | [[Image:Edinburgh-All-Actions.png|thumb|center|620px|'''Figure 1:''' Sugars are dark green squares, bonds between sugars are light green. From left to right are shown the actions of endoglucanase, exoglucanase, and β-glucosidase.]] |
==Simulating non-synergy vs. synergy== | ==Simulating non-synergy vs. synergy== | ||
| - | We can run two different simulations with the same settings. However, in one simulation the enzymes float about freely, whereas in the other they travel in triplets, each triplet containing one of each type of enzyme, side by side: | + | We can run two different simulations with the same settings. However, in one simulation the enzymes float about freely, whereas in the other they travel in triplets, each triplet containing one of each type of enzyme, side by side (this animation seems to work rather poorly in Safari): |
| - | + | <html> | |
| - | + | <div class="center"><div class="thumb tnone"><div class="thumbinner" style="width:622px;"><a href="/File:Syn-Variants-Animated.gif" class="image"><img alt="" src="/wiki/images/2/26/Syn-Variants-Animated.gif" width="620" height="210" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Syn-Variants-Animated.gif" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><b>Figure 2:</b> The enzymes' current positions are displayed as white, yellow, or blue squares. Note that in the synergistic system (right) they travel together. Endoglucanase is yellow; exoglucanase is white; β-glucosidase is blue. Exoglucanase is inhibited by nearby cellobiose.</div></div></div></div> | |
| + | </html> | ||
So, what happens if we run a large simulation? This: | So, what happens if we run a large simulation? This: | ||
| - | [[Image:Syn-v8-20-6000.png|thumb|center|620px|Iteration 6000 of a run with 20 copies of each enzyme per simulation. Dark regions are places where the cellulose has been degraded down to free glucose molecules. The left side has '''998''' free glucose molecules. The right (synergistic) side has '''2275'''.]] | + | [[Image:Syn-v8-20-6000.png|thumb|center|620px|'''Figure 3:''' Iteration 6000 of a run with 20 copies of each enzyme per simulation. Dark regions are places where the cellulose has been degraded down to free glucose molecules. The left side has '''998''' free glucose molecules. The right (synergistic) side has '''2275'''.]] |
| - | + | ==Animation== | |
| - | + | You can view an animation of a similar run to the above, [[Team:Edinburgh/Syn Animation | here]].<br>'''Warning:''' 1.5 MB file, has been known to hang certain browsers... | |
| - | + | ==Discussion== | |
| - | + | ||
| - | There are two ways to compile ' | + | This model is not intended as an accurate reflection of reality, but rather a simple demonstration that synergy works. The actual results seen depend heavily on the parameters passed to the program as [[#Command-line options|command-line arguments]]. Among the relevant settings are the number of enzymes, the size of the cellulose array, and the strength of cellobiose inhibition. |
| + | |||
| + | ===Baseline results=== | ||
| + | |||
| + | Some standard settings are used for parameters the user does not otherwise specify. These standard settings are fairly arbitrary, but they provide baseline results; we can then provide other parameters and see what difference they make. | ||
| + | |||
| + | We carried out 100 runs of the program with standard settings (20 of each enzyme per side, width and height 155, full inhibition of exoglucanase by nearby cellobiose, endoglucanase requiring at least 5 sugars on each side of a bond to cut it). | ||
| + | |||
| + | On average, it took the synergistic system '''7522''' iterations to liberate half the sugar (i.e. turn it into glucose monosaccharides), as opposed to '''13878''' for the non-synergistic system. | ||
| + | |||
| + | We can graph the progress of a typical run by counting the number of sugars present: | ||
| + | |||
| + | # As free glucose molecules, not bonded to anything. | ||
| + | # As one half of a cellobiose molecule. | ||
| + | # As part of a longer chain (cellulose). | ||
| + | |||
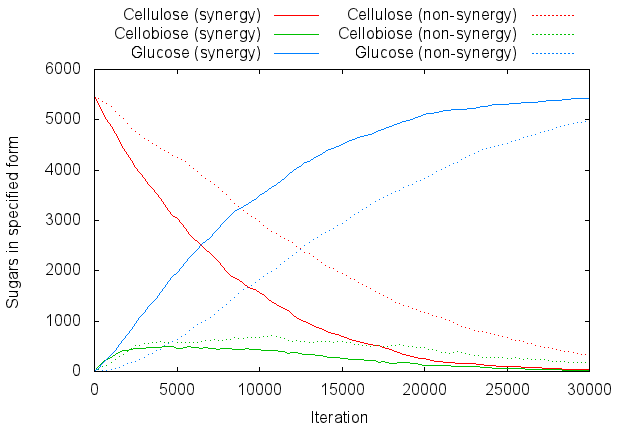
| + | For graphing purposes let us define cellulose as any chain with three or more sugars bonded together (meaning ''s-b-s-b-s'' is the smallest chain that counts as "cellulose"). Figure 4 is a graph of the number of sugar molecules present in each form: | ||
| + | |||
| + | [[Image:Edinburgh-Syn-Standard.png|thumb|center|620px| '''Figure 4:''' Comparison of the synergistic and non-synergistic systems, using the model's standard settings.]] | ||
| + | |||
| + | ===Effect of increased enzyme concentration=== | ||
| + | |||
| + | One obvious thing to try is increasing the enzyme concentration. Naturally, this will decrease the time taken to degrade the cellulose. But it also has a more subtle effect: since the enzymes are at higher density, they are automatically close to each other. Thus, even the model of free-floating enzymes has a sort of built-in synergy, and the result is rather similar to the explicitly synergistic model. | ||
| + | |||
| + | This is illustrated by Figure 5, which uses 500 enzymes of each type, instead of 20 '''(note carefully the different X axis):''' | ||
| + | |||
| + | [[Image:Edinburgh-Syn-500e.png|thumb|center|620px|'''Figure 5:''' 500 enzymes of each type are present. Time taken is drastically decreased, and the value of linking enzymes together is much less.]] | ||
| + | |||
| + | ===Effect of (no) cellobiose inhibition=== | ||
| + | |||
| + | Under standard settings, exoglucanase is completely inhibited if there is a nearby cellobiose molecule. This reflects the real-world fact that enzymes are generally inhibited by their own products. | ||
| + | |||
| + | We can disregard this assumption and see what happens (Figure 6). Since exoglucanase now works at higher efficiency, the amount of cellobiose that builds up is greater, but the time taken to degrade the cellulose is less. And since exoglucanase is no longer assisted by β-glucosidase, the value of synergy is less: | ||
| + | |||
| + | [[Image:Edinburgh-Syn-0inhib.png|thumb|center|620px|'''Figure 6:''' No inhibition of exoglucanase by cellobiose.]] | ||
| + | |||
| + | ==Conclusion== | ||
| + | |||
| + | Despite the simplicity of the model, it showed broadly similar results to the more complex [[Team:Edinburgh/Cellulases (Kappa model) | Kappa model]]. Both have shown that synergy actually makes sense! | ||
| + | |||
| + | ==Downloads== | ||
| + | |||
| + | * '''Source code:''' | ||
| + | :* [[:File:Syn.c.txt | syn.c]] | ||
| + | :* [[:File:Syn-Makefile.txt | Makefile]] | ||
| + | * '''Windows executable:''' | ||
| + | :* [[:File:Syn.zip | syn.zip]] | ||
| + | |||
| + | '''There are two ways to compile the code'''. The first is to just compile the C file as it stands, using a command like "gcc syn.c". When compiled in this way, there is no graphical display, except that the program saves .bmp graphics files every so often. | ||
However, if you have the SDL graphics library development files (called '''libsdl1.2-dev''' in Ubuntu), you can optionally download the Makefile and compile with "make". This version does not (by default) spit out any files; instead, it gives a graphical display of the action as it occurs. | However, if you have the SDL graphics library development files (called '''libsdl1.2-dev''' in Ubuntu), you can optionally download the Makefile and compile with "make". This version does not (by default) spit out any files; instead, it gives a graphical display of the action as it occurs. | ||
| Line 56: | Line 111: | ||
Note that, to work, the Makefile should be called "Makefile" and the C file should be called "syn.c". | Note that, to work, the Makefile should be called "Makefile" and the C file should be called "syn.c". | ||
| - | + | ===Command-line options=== | |
| - | == | + | The following command-line options should work. Default values are shown where applicable. (The default ''endpoint'' and ''bmptick'' depend on whether SDL support is compiled in.) |
| + | |||
| + | ====Basic settings==== | ||
| + | |||
| + | : '''--width''' ''155'' — width of each simulation | ||
| + | : '''--height''' ''155'' — height of each simulation | ||
| + | : '''--margin''' ''4'' — size of the margin (region of no cellulose) | ||
| + | : '''--enzymes''' ''20'' — number of enzymes of each type per simulation | ||
| + | : '''--endpoint''' ''x'' — how many iterations to run (0 for unlimited) | ||
| + | : '''--seed''' ''x'' — use specified random seed (results may still differ across platforms) | ||
| + | |||
| + | ====Cellobiose inhibition of exoglucanase==== | ||
| + | |||
| + | : '''--bioseinhib''' ''1.0'' — strength of cellobiose inhibition of exoglucanase, from 0 to 1 | ||
| + | : '''--bioseinhibleft''' ''1.0'' — as above, but sets value for left simulation only | ||
| + | : '''--bioseinhibright''' ''1.0'' — as above, but sets value for right simulation only | ||
| + | |||
| + | ====Endoglucanase behaviour==== | ||
| + | |||
| + | : '''--endoreq''' ''5'' — how close to the end of a chain endoglucanase can cut | ||
| + | : '''--endoreqleft''' ''5'' — as above, but sets value for left simulation only | ||
| + | : '''--endoreqright''' ''5'' — as above, but sets value for right simulation only | ||
| + | |||
| + | ====Synergy (enzymes physically linked)==== | ||
| + | |||
| + | : '''--nosyn''' — do not use synergy on either side | ||
| + | : '''--bothsyn''' — use synergy on both sides | ||
| + | |||
| + | ====Display and reporting==== | ||
| - | + | : '''--bmptick''' ''x'' — how often to spit out .bmp graphics files (0 for never) | |
| + | : '''--bmpmag''' ''1'' — magnification of .bmp files | ||
| + | : '''--sdltick''' ''1'' — how often to draw to the SDL window (1 to draw every frame) | ||
| + | : '''--sdlmag''' ''2'' — magnification of SDL window | ||
| + | : '''--delay''' ''20'' — milliseconds to delay after drawing to SDL window; some machines require this | ||
| + | : '''--reporttick''' ''250'' — how often to print count of glucose and cellobiose to the console (0 for never) | ||
==References== | ==References== | ||
Latest revision as of 19:10, 16 October 2011
Cellulases (C model)
As explained in the abstract, the feasibility of our improved biorefineries depends on exploiting synergy between enzymes. Therefore, we attempted to use the C programming language to construct a model which could prove that synergy is, or might be, a real phenomenon affecting cellulase action.
Contents |
Synergy explained
It can seem almost magical that having enzymes closer together can increase their usefulness, but consider the following statements about cellulose degradation, all of which are true ([http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-091208-085603 Fontes and Gilbert, 2010]):
- Exoglucanase chews away at the end of a cellulose chain, producing cellobiose sugars.
- Endoglucanase cuts cellulose chains in the middle, essentially turning one chain into two.
- Cellobiose inhibits the action of the above enzymes.
- β-glucosidase cuts cellobiose in half, producing two glucose molecules.
Facts 1 and 2 mean that exoglucanase works best with endoglucanase nearby, since every time endoglucanase acts, it produces new chain-ends for exoglucanase to attack. Meanwhile, facts 1, 3, and 4 mean that β-glucosidase helps nearby copies of the other enzymes by preventing the cellobiose from inhibiting them.
Syn: a simple demonstration of synergy
We can demonstrate that there is (potentially) a huge difference between the synergistic and non-synergistic systems with Edinburgh's Syn program, written in C. This is a 2D simulation that works in the following way:
- The world is a 2D grid.
- Each spot in the grid can contain a sugar, a bond between sugars, or nothing.
- The world begins with an array of cellulose.
- Cellulose is modelled as horizontal chains with alternating sugars and bonds: s-b-s-b-s-b-s-b-s-b-s, etc.
- There are three enzyme types: endoglucanase, exoglucanase, and β-glucosidase.
- The enzymes move about randomly in manner similar to Brownian motion.
- If an enzyme is in the same place as a bond, it can destroy it:
- Endoglucanase only cuts bonds away from the ends of a chain (some minimum distance is specified).
- Exoglucanase only cuts bonds if this results in a cellobiose molecule (s-b-s) forming.
- β-glucosidase only cuts cellobiose bonds.
- There is inhibition of exoglucanase by cellobiose; it does not cut bonds (or has a reduced chance to do so) if there is a nearby cellobiose molecule.
So, the actions of the enzymes are as follows:
Simulating non-synergy vs. synergy
We can run two different simulations with the same settings. However, in one simulation the enzymes float about freely, whereas in the other they travel in triplets, each triplet containing one of each type of enzyme, side by side (this animation seems to work rather poorly in Safari):
So, what happens if we run a large simulation? This:
Animation
You can view an animation of a similar run to the above, here.
Warning: 1.5 MB file, has been known to hang certain browsers...
Discussion
This model is not intended as an accurate reflection of reality, but rather a simple demonstration that synergy works. The actual results seen depend heavily on the parameters passed to the program as command-line arguments. Among the relevant settings are the number of enzymes, the size of the cellulose array, and the strength of cellobiose inhibition.
Baseline results
Some standard settings are used for parameters the user does not otherwise specify. These standard settings are fairly arbitrary, but they provide baseline results; we can then provide other parameters and see what difference they make.
We carried out 100 runs of the program with standard settings (20 of each enzyme per side, width and height 155, full inhibition of exoglucanase by nearby cellobiose, endoglucanase requiring at least 5 sugars on each side of a bond to cut it).
On average, it took the synergistic system 7522 iterations to liberate half the sugar (i.e. turn it into glucose monosaccharides), as opposed to 13878 for the non-synergistic system.
We can graph the progress of a typical run by counting the number of sugars present:
- As free glucose molecules, not bonded to anything.
- As one half of a cellobiose molecule.
- As part of a longer chain (cellulose).
For graphing purposes let us define cellulose as any chain with three or more sugars bonded together (meaning s-b-s-b-s is the smallest chain that counts as "cellulose"). Figure 4 is a graph of the number of sugar molecules present in each form:
Effect of increased enzyme concentration
One obvious thing to try is increasing the enzyme concentration. Naturally, this will decrease the time taken to degrade the cellulose. But it also has a more subtle effect: since the enzymes are at higher density, they are automatically close to each other. Thus, even the model of free-floating enzymes has a sort of built-in synergy, and the result is rather similar to the explicitly synergistic model.
This is illustrated by Figure 5, which uses 500 enzymes of each type, instead of 20 (note carefully the different X axis):
Effect of (no) cellobiose inhibition
Under standard settings, exoglucanase is completely inhibited if there is a nearby cellobiose molecule. This reflects the real-world fact that enzymes are generally inhibited by their own products.
We can disregard this assumption and see what happens (Figure 6). Since exoglucanase now works at higher efficiency, the amount of cellobiose that builds up is greater, but the time taken to degrade the cellulose is less. And since exoglucanase is no longer assisted by β-glucosidase, the value of synergy is less:
Conclusion
Despite the simplicity of the model, it showed broadly similar results to the more complex Kappa model. Both have shown that synergy actually makes sense!
Downloads
- Source code:
- Windows executable:
There are two ways to compile the code. The first is to just compile the C file as it stands, using a command like "gcc syn.c". When compiled in this way, there is no graphical display, except that the program saves .bmp graphics files every so often.
However, if you have the SDL graphics library development files (called libsdl1.2-dev in Ubuntu), you can optionally download the Makefile and compile with "make". This version does not (by default) spit out any files; instead, it gives a graphical display of the action as it occurs.
Note that, to work, the Makefile should be called "Makefile" and the C file should be called "syn.c".
Command-line options
The following command-line options should work. Default values are shown where applicable. (The default endpoint and bmptick depend on whether SDL support is compiled in.)
Basic settings
- --width 155 — width of each simulation
- --height 155 — height of each simulation
- --margin 4 — size of the margin (region of no cellulose)
- --enzymes 20 — number of enzymes of each type per simulation
- --endpoint x — how many iterations to run (0 for unlimited)
- --seed x — use specified random seed (results may still differ across platforms)
Cellobiose inhibition of exoglucanase
- --bioseinhib 1.0 — strength of cellobiose inhibition of exoglucanase, from 0 to 1
- --bioseinhibleft 1.0 — as above, but sets value for left simulation only
- --bioseinhibright 1.0 — as above, but sets value for right simulation only
Endoglucanase behaviour
- --endoreq 5 — how close to the end of a chain endoglucanase can cut
- --endoreqleft 5 — as above, but sets value for left simulation only
- --endoreqright 5 — as above, but sets value for right simulation only
Synergy (enzymes physically linked)
- --nosyn — do not use synergy on either side
- --bothsyn — use synergy on both sides
Display and reporting
- --bmptick x — how often to spit out .bmp graphics files (0 for never)
- --bmpmag 1 — magnification of .bmp files
- --sdltick 1 — how often to draw to the SDL window (1 to draw every frame)
- --sdlmag 2 — magnification of SDL window
- --delay 20 — milliseconds to delay after drawing to SDL window; some machines require this
- --reporttick 250 — how often to print count of glucose and cellobiose to the console (0 for never)
References
- Fontes CMGA, Gilbert HJ (2010) [http://www.annualreviews.org/doi/abs/10.1146/annurev-biochem-091208-085603 Cellulosomes: highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates]. Annual Review of Biochemistry 79: 655-81 (doi: 10.1146/annurev-biochem-091208-085603).
 "
"