Team:Wageningen UR/Project/IntroductionProj1
From 2011.igem.org
(→The Synchroscillator: a Synchronized Oscillatory System) |
|||
| (18 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <head> | ||
| + | <style type="text/css"> | ||
| + | |||
| + | ul li a.currentlink1 { | ||
| + | color: black !important; | ||
| + | } | ||
| + | |||
| + | ul li a.currentlinktop3 { | ||
| + | color: #63a015 !important; | ||
| + | } | ||
| + | |||
| + | ul li a.currentlinktop4 { | ||
| + | color: black !important; | ||
| + | display: block; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | </head> | ||
| + | <bod> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
{{:Team:Wageningen_UR/Templates/Header}} | {{:Team:Wageningen_UR/Templates/Header}} | ||
{{:Team:Wageningen_UR/Templates/NavigationTop1}} | {{:Team:Wageningen_UR/Templates/NavigationTop1}} | ||
| - | == Synchronized Oscillatory System == | + | == The Synchroscillator: a Synchronized Oscillatory System == |
{{:Team:Wageningen_UR/Templates/NavigationLeft}} | {{:Team:Wageningen_UR/Templates/NavigationLeft}} | ||
| - | {{:Team:Wageningen_UR/Templates/Style | text= | + | {{:Team:Wageningen_UR/Templates/Style | text= __NOTOC__ |
| - | + | ||
| - | + | ||
| - | + | === Abstract === | |
| + | One aim of synthetic biology is to re-engineer naturally occurring gene circuits to produce artificial systems that behave predictably. Our project involved providing additional functionality and also streamlining a recently published synchronized oscillatory circuit in an attempt to reproduce and further characterize its dynamics. Our genetic circuit consists of modified (and BioBricked) elements of the ''Vibrio fischeri'' lux quorum sensing system composed to form interconnected positive and negative feedback loops, which dynamically regulate the expression of GFP. In order to provide our ''E. coli'' host with the right environment required for population-wide oscillations, we designed and manufactured a custom [[Team:Wageningen_UR/Project/Devices| flow-chamber]] capable of maintaining a defined cell population while independently varying the growth conditions. The chamber was specifically designed for time-lapse studies with a fluorescence microscope. We detected synchronized oscillatory gene expression under zero-flow conditions, suggesting an unexpected level of robustness. This should facilitate its integration with more advanced genetic circuits. | ||
| + | [[Team:Wageningen_UR/Project/CompleteProject1Description| Read more]] | ||
| - | |||
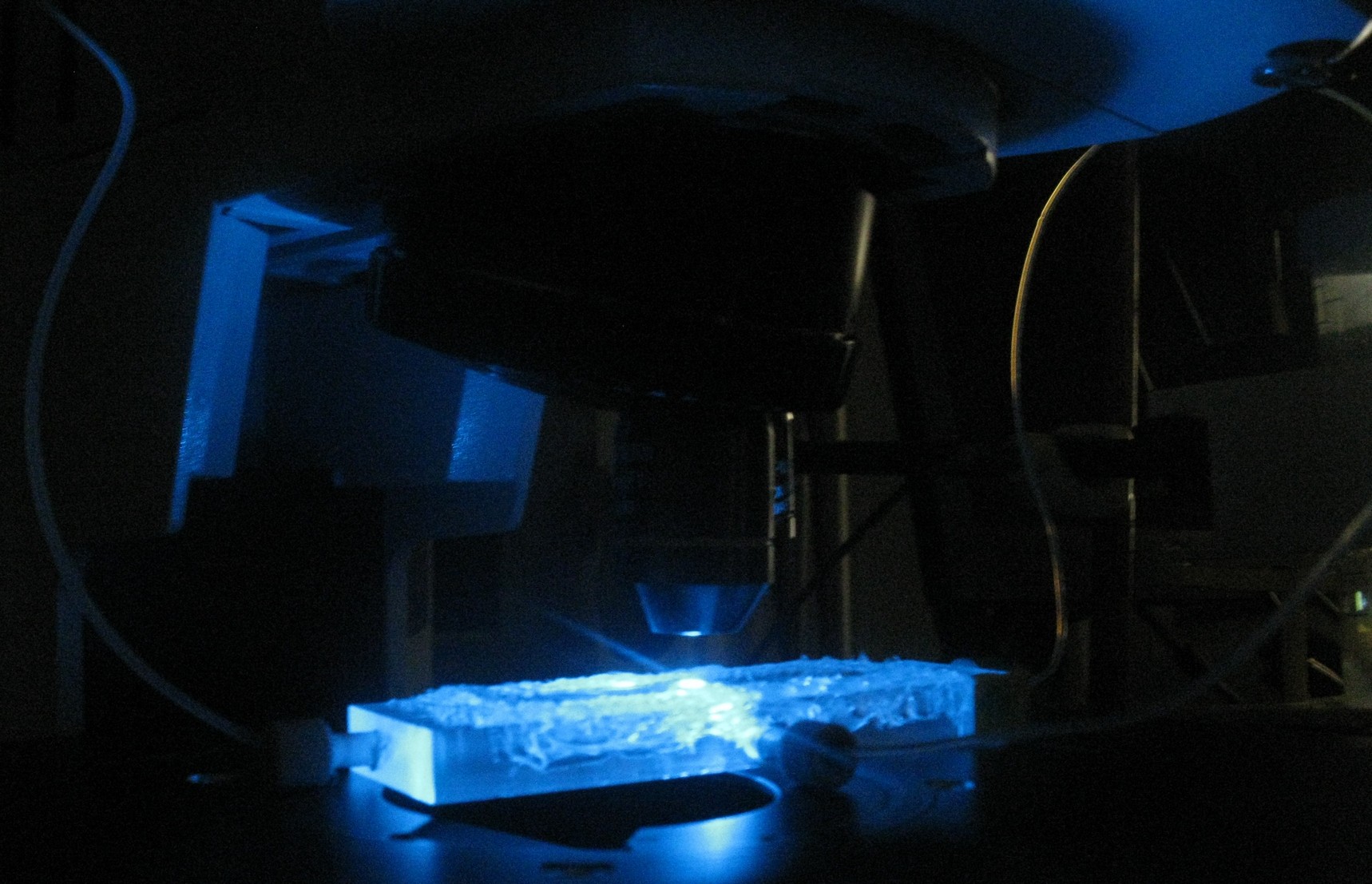
| + | [[File:Measuring_GFP_WUR.jpg|600px|center]] | ||
| + | '''Fig. 1''' ''Olympus microscope exciting GFP in the flow chamber, the Laboratory of Microbiology, Wageningen'' | ||
}} | }} | ||
Latest revision as of 00:21, 22 September 2011
 "
"