Team:British Columbia/Notebook/Week 9
From 2011.igem.org
(→Characterization: Limonene Synthase (Lims1) from registry: BBa_K118025) |
|||
| (13 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Template:Notebook}} | {{Template:Notebook}} | ||
| - | <b> Week 9: July 31-August 6</b> | + | <html> |
| + | <style> | ||
| + | #bod {width:935px; float:left; background-color: white; margin-left: 15px; margin-top:10px;}</style> | ||
| + | <div id="bod"><b> Week 9: July 31-August 6</b> <html><a name="w9"></a></html> | ||
==Lab Meeting - August 2== | ==Lab Meeting - August 2== | ||
| Line 17: | Line 20: | ||
Joe is troubleshooting his PCR reaction in preparation of his yeast and biobrick plasmid construction. This time he used an annealing temperature of 60 degree Celsius, the low range for Phusion Hot Start Polymerase. The results after gel verification show a supercoiled DNA above the 1kb ladder and a small non-specific high molecular weight smear near 6-8kb. This was unexpected as the pinene synathase should be around 2.1kb long. It is unknown why this may be. Next time, he will try increasing the extension time and a higher annealing temperature. | Joe is troubleshooting his PCR reaction in preparation of his yeast and biobrick plasmid construction. This time he used an annealing temperature of 60 degree Celsius, the low range for Phusion Hot Start Polymerase. The results after gel verification show a supercoiled DNA above the 1kb ladder and a small non-specific high molecular weight smear near 6-8kb. This was unexpected as the pinene synathase should be around 2.1kb long. It is unknown why this may be. Next time, he will try increasing the extension time and a higher annealing temperature. | ||
| - | Yup... troubleshooting the previous PCR reaction again. Joe used an annealing temperature of 63 degree Celsius and 65 degree Celsius, plus a 2 minute extension time. The same results were seen after the gel verification. However, a clear band could be seen this time at 7-8kb. Again, this is not the product Joe wanted. It should be noted that the synthase gene has a long stretch A's at the 3' end of the gene. Therefore, the reverse primer has a long stretch of T's. It may be the forward primer is annealing to the sequence while the reverse primer is not annealing due to the long stretch of the same nucleotide. | + | Yup... troubleshooting the previous PCR reaction again. Joe used an annealing temperature of 63 degree Celsius and 65 degree Celsius, plus a 2 minute extension time. The same results were seen after the gel verification. However, a clear band could be seen this time at 7-8kb. Again, this is not the product Joe wanted. It should be noted that the synthase gene has a long stretch A's at the 3' end of the gene. Therefore, the reverse primer has a long stretch of T's. It may be the forward primer is annealing to the sequence while the reverse primer is not annealing due to the long stretch of the same nucleotide. This is enough...he is reordering new primers. |
==1,8-Cineole== | ==1,8-Cineole== | ||
| - | Good news and bad news. The good news is that the PCR to put restriction sites on the end of the synthases was a resounding success. The bad news is that a restriction digest with just the genes of the synthases (as opposed to the plasmid with the genes) showed that pg-tps-1,8-cineole still had a XBAI site, and pgxe-tps-1,8-cineole still had a PSTI site. Because of this another round of SDMs was needed. | + | Good news and bad news. The good news is that the PCR to put restriction sites on the end of the synthases was a resounding success. The bad news is that a restriction digest with just the genes of the synthases (as opposed to the plasmid with the genes) showed that pg-tps-1,8-cineole synthase still had a XBAI site, and pgxe-tps-1,8-cineole synthase still had a PSTI site. Because of this another round of SDMs was needed. |
The SDMs worked well, and now the genes should be free from the illegal restriction sites. The team has yet to have any success with sequencing, which could easily confirm this. | The SDMs worked well, and now the genes should be free from the illegal restriction sites. The team has yet to have any success with sequencing, which could easily confirm this. | ||
| - | In the mean time, Jacob tried putting his semi-SDM'd | + | In the mean time, Jacob tried putting his semi-SDM'd synthase genes as well as the original synthase genes into the 415 PAG GPD and GAL shuttle vectors. The original synthase genes could confirm that the SDMs did not change the function of the gene, and the semi-SDM'd genes on the PAG 415 could undergo another round of SDM as easily as on the PET plasmids, and the ligation would already be done. However, after growing them up, overnight culturing, miniprepping, and digesting, it became clear that the ligation was not successful for any synthase. A ~4 kb product was produced for both the cut and uncut plasmids, while the 1,8-cineole synthase genes are about 1.7 kb, the 415 PAG GPD and GAL are a little over 8 kb, and Jacob cut only about 1.2 kb (the CCDB gene and a little more out of the shuttle vector). Perplexing results! |
==beta-Pinene== | ==beta-Pinene== | ||
| Line 45: | Line 48: | ||
Success! Gel verification of the one HMG2 and two IDI1 PCR product showed bands at 4kb and 1.5 kb, respectively. Nanodrop measurements on purified product showed DNA levels of ~30 ng/uL for HMG2 and ~150ng/uL for IDI1. | Success! Gel verification of the one HMG2 and two IDI1 PCR product showed bands at 4kb and 1.5 kb, respectively. Nanodrop measurements on purified product showed DNA levels of ~30 ng/uL for HMG2 and ~150ng/uL for IDI1. | ||
| + | ==ERG20 & erg20-2== | ||
| - | ==Characterization: Limonene Synthase ( | + | Gurpal started by verifying that his PCR from last week was a success. He expected his ERG20 and erg20-2 genes to be about 1070 bps. His gel image verified that his PCR was a success! |
| + | |||
| + | Here is the gel image! | ||
| + | [[File:Gel_-_PCR_Product_Start_ERG_biobricks.jpg | thumb | center |]] | ||
| + | |||
| + | Next, he tried to perform a restriction digest to prepare for ligating his insert genes with a bio brick vector. Unfortunately, the unexpected happened and his PCR digest products completely vaporized in the PCR machine. Only a few tears were shed. | ||
| + | |||
| + | ==Characterization: Limonene Synthase (LIMS1) from registry: BBa_K118025== | ||
[[File:smilietube.jpg | thumb | left | 95px | Smiley face cells aka C41 DE3 cells]] | [[File:smilietube.jpg | thumb | left | 95px | Smiley face cells aka C41 DE3 cells]] | ||
| Line 54: | Line 65: | ||
Daisy checked on the plates and there were no transformants. The transformation did not work. Daisy thinks it may be because she did not let the cells incubate long enough. She consulted Chris Keeling and his colleagues. They mentioned it may be better to transform into DH5 alpha first and then miniprep the plasmids to transform into C41 DE3. | Daisy checked on the plates and there were no transformants. The transformation did not work. Daisy thinks it may be because she did not let the cells incubate long enough. She consulted Chris Keeling and his colleagues. They mentioned it may be better to transform into DH5 alpha first and then miniprep the plasmids to transform into C41 DE3. | ||
| + | ==Characterization: Limonene Synthase (LIMS1) from registry: BBa_I742110== | ||
For further characterization, Vicki is going to design primers to take out the (+)-limonene synthase from the composite part BBa_K118025. The primers will contain restriction enzyme sites SpeI and XhoI. Her objective is to get the synthase onto a yeast plasmid so she can overexpress the product, then the expression level in bacteria and in yeast will be compared. | For further characterization, Vicki is going to design primers to take out the (+)-limonene synthase from the composite part BBa_K118025. The primers will contain restriction enzyme sites SpeI and XhoI. Her objective is to get the synthase onto a yeast plasmid so she can overexpress the product, then the expression level in bacteria and in yeast will be compared. | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <a href="https://2011.igem.org/Team:British_Columbia/Notebook"><center><b>Back to the Notebook</b></center></a> | ||
Latest revision as of 23:14, 16 October 2011

 |
 |
 |
 |
 |
Contents |
Lab Meeting - August 2
Chris Keeling came to the meeting and briefly outlined the protocol for preparation of samples for GC-MS (gas-chromatography mass-spectroscopy). He told us to speak to Neils in the Ellis Lab for a more detailed protocol.
The Wiki
Vicki feels that Dreamweaver is too specialized (ie. has Dreamweaver specific codes for menus, etc.) such that it is difficult to transfer the code over to the wiki. She realizes that it is much simpler to write the code herself. Vicki and Alina begin re-formatting the wiki. Let the trial and errors begin. Slow and steady wins the race, right?
Vicki, Alina, and Joanne Fox met up during the week to discuss the format; the goal: a clean, slick, easy to navigate, intuitive, and simple wiki. We felt very excited about the re-design. Both Vicki and Alina went home to slave away at coding, and creating images with photoshop, respectively.
A few (almost) sleepless nights, the result is what you see before you.
alpha-Pinene
Joe is troubleshooting his PCR reaction in preparation of his yeast and biobrick plasmid construction. This time he used an annealing temperature of 60 degree Celsius, the low range for Phusion Hot Start Polymerase. The results after gel verification show a supercoiled DNA above the 1kb ladder and a small non-specific high molecular weight smear near 6-8kb. This was unexpected as the pinene synathase should be around 2.1kb long. It is unknown why this may be. Next time, he will try increasing the extension time and a higher annealing temperature.
Yup... troubleshooting the previous PCR reaction again. Joe used an annealing temperature of 63 degree Celsius and 65 degree Celsius, plus a 2 minute extension time. The same results were seen after the gel verification. However, a clear band could be seen this time at 7-8kb. Again, this is not the product Joe wanted. It should be noted that the synthase gene has a long stretch A's at the 3' end of the gene. Therefore, the reverse primer has a long stretch of T's. It may be the forward primer is annealing to the sequence while the reverse primer is not annealing due to the long stretch of the same nucleotide. This is enough...he is reordering new primers.
1,8-Cineole
Good news and bad news. The good news is that the PCR to put restriction sites on the end of the synthases was a resounding success. The bad news is that a restriction digest with just the genes of the synthases (as opposed to the plasmid with the genes) showed that pg-tps-1,8-cineole synthase still had a XBAI site, and pgxe-tps-1,8-cineole synthase still had a PSTI site. Because of this another round of SDMs was needed.
The SDMs worked well, and now the genes should be free from the illegal restriction sites. The team has yet to have any success with sequencing, which could easily confirm this.
In the mean time, Jacob tried putting his semi-SDM'd synthase genes as well as the original synthase genes into the 415 PAG GPD and GAL shuttle vectors. The original synthase genes could confirm that the SDMs did not change the function of the gene, and the semi-SDM'd genes on the PAG 415 could undergo another round of SDM as easily as on the PET plasmids, and the ligation would already be done. However, after growing them up, overnight culturing, miniprepping, and digesting, it became clear that the ligation was not successful for any synthase. A ~4 kb product was produced for both the cut and uncut plasmids, while the 1,8-cineole synthase genes are about 1.7 kb, the 415 PAG GPD and GAL are a little over 8 kb, and Jacob cut only about 1.2 kb (the CCDB gene and a little more out of the shuttle vector). Perplexing results!
beta-Pinene
5 colonies grew on Marianne's biobrick plate! Marianne ran a colony PCR on all the colonies and conducted a gel verification. Unfortunately, none of them showed bands at the correct length (~2000bp). It seems that the miniprepped plasmids of pSB1C3 we have are problematic. Therefore, the plan is to resuspend the pSB1C3 in the 2011 Spring Biobrick Kit Plate 1 A3, transform them, and miniprep them for a fresh tube of pSB1C3.
While Marianne is working on getting a biobrick part, Vicki is helping her make more yeast plasmids. What Marianne already has: SDMed pin2 synthase on both GAL and GPD promoter. And what Vicki is working on getting: Original (non-SDM) with a his-tag on GPD promoter, original (non-SDM) without a his-tag on GPD promoter, SDMed with a his-tag on GPD promoter. However, before Vicki can begin on assembling these constructs, she needs to mini-prep more of the yeast plasmid (pAD415GPD) that contains GPD promoter and Leu marker. After, she digested and ligated the yeast plasmid and the different synthases (original+his, original+no-his, SDM-his-tag); then, Vicki transformed them into E.coli with a positive and negative control. Results pending!
IDI1, HMG2 metabolic genes
Sam performed PCR again on the two IDI1 plasmid samples and the HMG2 plasmid sample. He used twice the amount of primer (8.0uL of 10uM forward and reverse, each) per 50uL reaction and did two reactions for each sample, one with 0.5 uL 50 mM MgCl2 and one without. The annealing temperature was raised to 58 oC from 57 oC.
Success! Gel verification of the one HMG2 and two IDI1 PCR product showed bands at 4kb and 1.5 kb, respectively. Nanodrop measurements on purified product showed DNA levels of ~30 ng/uL for HMG2 and ~150ng/uL for IDI1.
ERG20 & erg20-2
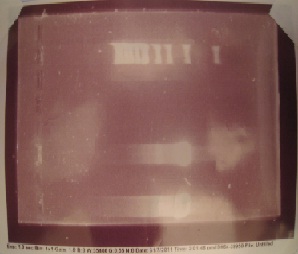
Gurpal started by verifying that his PCR from last week was a success. He expected his ERG20 and erg20-2 genes to be about 1070 bps. His gel image verified that his PCR was a success!
Here is the gel image!
Next, he tried to perform a restriction digest to prepare for ligating his insert genes with a bio brick vector. Unfortunately, the unexpected happened and his PCR digest products completely vaporized in the PCR machine. Only a few tears were shed.
Characterization: Limonene Synthase (LIMS1) from registry: BBa_K118025
Daisy got a tube of C41 DE3 expression cells (also called smiley face cells) from Chris Keeling. She is going to use these cells to transform the plasmid with the limonene synthase into the cells. These cells are specially designed to express difficult-to-express proteins. Chris Keeling has often mentioned that synthases tend to be difficult to express.
Daisy checked on the plates and there were no transformants. The transformation did not work. Daisy thinks it may be because she did not let the cells incubate long enough. She consulted Chris Keeling and his colleagues. They mentioned it may be better to transform into DH5 alpha first and then miniprep the plasmids to transform into C41 DE3.
Characterization: Limonene Synthase (LIMS1) from registry: BBa_I742110
For further characterization, Vicki is going to design primers to take out the (+)-limonene synthase from the composite part BBa_K118025. The primers will contain restriction enzyme sites SpeI and XhoI. Her objective is to get the synthase onto a yeast plasmid so she can overexpress the product, then the expression level in bacteria and in yeast will be compared.
 "
"