Team:NTNU Trondheim
From 2011.igem.org
(Difference between revisions)
(→Project description) |
|||
| (64 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| + | =Introduction= | ||
| - | |||
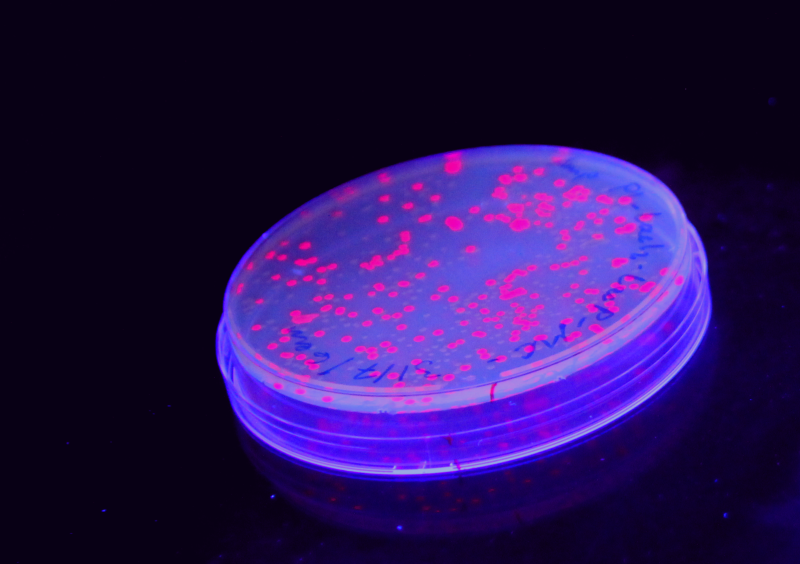
iGEM is the premiere international competition in Synthetic Biology for university students, this year with more than 160 teams from all over the world. For the first time, a Norwegian team is participating! Our team, consisting of students from the Norwegian University of Science and Technology (NTNU) are really excited to be able to compete, and we are very much looking forward to honing our skills in synthetic biology. Our project idea is based on using the alarmone [http://en.wikipedia.org/wiki/Guanosine_pentaphosphate ppGpp] to detect stress in ''E. coli'', and basically make it turn red with anger! | iGEM is the premiere international competition in Synthetic Biology for university students, this year with more than 160 teams from all over the world. For the first time, a Norwegian team is participating! Our team, consisting of students from the Norwegian University of Science and Technology (NTNU) are really excited to be able to compete, and we are very much looking forward to honing our skills in synthetic biology. Our project idea is based on using the alarmone [http://en.wikipedia.org/wiki/Guanosine_pentaphosphate ppGpp] to detect stress in ''E. coli'', and basically make it turn red with anger! | ||
| - | = | + | {|style="font-size: 20px;" |
| + | |- | ||
| + | |[[:Team:NTNU_Trondheim/Data|Our Data page]] | ||
| + | |} | ||
| + | {| | ||
| + | |- | ||
| + | |[[File:NTNU2011 forside s.png|550px]] | ||
| + | |{{:Team:NTNU_Trondheim/Judging}} | ||
| + | |} | ||
| - | |||
| - | + | {|align="center" | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | {|align=" | + | |
|<html> | |<html> | ||
| - | <div id="fb-root"></div><script src="http://connect.facebook.net/en_US/all.js#xfbml=1"></script><fb:like-box href="http://www.facebook.com/pages/iGEM-NTNU-2011/125109070907385" width="292" show_faces="false" border_color="" stream="false" header="true"></fb:like-box> | + | <div id="fb-root"></div> |
| + | <script src="http://connect.facebook.net/en_US/all.js#xfbml=1"></script><fb:like-box href="http://www.facebook.com/pages/iGEM-NTNU-2011/125109070907385" width="292" show_faces="false" border_color="" stream="false" header="true"></fb:like-box> | ||
</html> | </html> | ||
|} | |} | ||
| - | + | {{:Team:NTNU_Trondheim/NTNU_footer}} | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
Latest revision as of 07:34, 21 September 2011

Introduction
iGEM is the premiere international competition in Synthetic Biology for university students, this year with more than 160 teams from all over the world. For the first time, a Norwegian team is participating! Our team, consisting of students from the Norwegian University of Science and Technology (NTNU) are really excited to be able to compete, and we are very much looking forward to honing our skills in synthetic biology. Our project idea is based on using the alarmone [http://en.wikipedia.org/wiki/Guanosine_pentaphosphate ppGpp] to detect stress in E. coli, and basically make it turn red with anger!
| Our Data page |
|
|
 "
"