Team:Freiburg/Notebook/27 July
From 2011.igem.org
(Difference between revisions)
(→Gel) |
|||
| (19 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{:Team:Freiburg/Templates/header}} | ||
| + | <html> | ||
| + | <div id="notebook-page-header"> | ||
| + | <div id="notebook-back" width="100px" > | ||
| + | <a href="https://2011.igem.org/Team:Freiburg/Notebook/26_July">Previous entry</a> | ||
| + | </div> | ||
| + | <div id="notebook-title"> | ||
| + | <a href="https://2011.igem.org/Team:Freiburg/Notebook"> 27 July </a> | ||
| + | </div> | ||
| + | <div id="notebook-next"> | ||
| + | <a href="https://2011.igem.org/Team:Freiburg/Notebook/28_July">Next entry</a> | ||
| + | </div> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
==Commons== | ==Commons== | ||
===Gel=== | ===Gel=== | ||
| Line 6: | Line 21: | ||
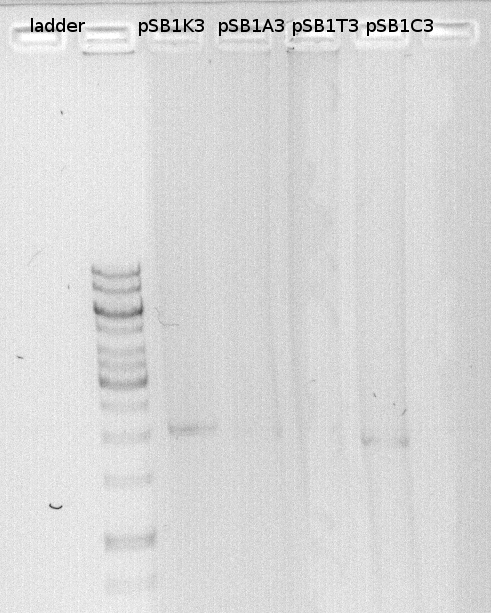
We loaded the PCR products of the vectors on a gel and measured the DNA concentration. DNA concentration were high and we diluted the vectors to get an endconcentration of 25ng/microl. | We loaded the PCR products of the vectors on a gel and measured the DNA concentration. DNA concentration were high and we diluted the vectors to get an endconcentration of 25ng/microl. | ||
| - | [[File:.Gelbild_Vektoren_27_07.jpg]] | + | '''DNA-concentration measured with nanodrop:'''<br/> |
| + | {|cellpadding="10" cellspacing="0" border="1" | ||
| + | |sample | ||
| + | |DNA concentration (ng/μl) | ||
| + | |- | ||
| + | |pSB1C3 | ||
| + | |132.1 | ||
| + | |- | ||
| + | |pSB1K3 | ||
| + | |101.5 | ||
| + | |- | ||
| + | |pSB1A3 | ||
| + | |83.9 | ||
| + | |- | ||
| + | |pSB1T3 | ||
| + | |116.2 | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[File:.Gelbild_Vektoren_27_07.jpg|350px|caption]] | ||
==<span style="color:green;">green light receptor</span>== | ==<span style="color:green;">green light receptor</span>== | ||
| - | === | + | ===Results of Sequencing=== |
| + | |||
| + | '''Investigators:Julia''' | ||
| + | |||
| + | |||
| + | CcaS 4a2 seemed to miss about 20bp, CcaS 4b2 is correct. | ||
| + | |||
| + | ===Quickchange PCR of CcaS and CcaR=== | ||
| + | |||
| + | '''Investigators:Julia''' | ||
| + | |||
| + | in order to eliminate iGEM restriction sites of EcoRI within CcaS and CcaR, we designed mutagenesis primer and performed a quickchange PCR. | ||
| + | |||
| + | ===Protocol=== | ||
| + | |||
| + | total reaction volume is 50µl: | ||
| + | |||
| + | 32,5 µl H2O<br/> | ||
| + | 10 µl Phusion Buffer 5x<br/> | ||
| + | 2,5 µl Primer up<br/> | ||
| + | 2,5 µl Primer dw<br/> | ||
| + | 1 µl DNA template, concentration has to be 5ng/µl<br/> | ||
| + | 0,5 µl Phusion<br/> | ||
| - | + | Primer for CcaR:<br/> | |
| + | ccaR_mut1_up: CCtcactaATGAGgATcCTTTTAGTGGAGG<br/> | ||
| + | ccaR_mut1_dw:CCTCCACTAAAAGgATcCTCATtagtgaGG<br/> | ||
| + | <br/> | ||
| + | Primer for CcaS:<br/> | ||
| + | ccaS_mut1_up:CCGGGCCGAGgATcCTCTATGTCAATG<br/> | ||
| + | ccaS_mut1_dw:CATTGACATAGAGgATcCTCGGCCCGG<br/> | ||
| + | <br/> | ||
| + | Primer for CcaS:<br/> | ||
| + | ccaS_mut2_up:GGACCAAAAACgAGTCGCACTGAATTAG<br/> | ||
| + | ccaS_mut2_dw:CTAATTCAGTGCGACTcGTTTTTGGTCC<br/> | ||
==<span style="color:blue;">blue light receptor</span>== | ==<span style="color:blue;">blue light receptor</span>== | ||
| Line 22: | Line 88: | ||
We repeated the PCR again, with higher temperatures this time. | We repeated the PCR again, with higher temperatures this time. | ||
After PCR we digested with DpnI 37°C for 1 hour and then purified the DNA with PCR purifictaion kit. Afterwards we measured the DNA concentration and loaded 2 microl to a gel. | After PCR we digested with DpnI 37°C for 1 hour and then purified the DNA with PCR purifictaion kit. Afterwards we measured the DNA concentration and loaded 2 microl to a gel. | ||
| + | |||
'''PCR''' | '''PCR''' | ||
| Line 33: | Line 100: | ||
|- | |- | ||
| - | | colspan="2" style="border:0.0069in solid #00000a;padding-top:0in;padding-bottom:0in;padding-left:0.075in;padding-right:0.075in;"| Continue from Experiment: NOT-Gate and LovTAP with Gibson-overhangs (now we use a little bit higher temperature for primer annealing) (Date): 25.07.11 | + | | colspan="2" style="border:0.0069in solid #00000a;padding-top:0in;padding-bottom:0in;padding-left:0.075in;padding-right:0.075in;"| Continue from Experiment: NOT-Gate and LovTAP with Gibson-overhangs (now we use a little bit higher temperature for primer annealing) -(Date): 25.07.11 |
(Name): Sandra, Sophie | (Name): Sandra, Sophie | ||
| Line 90: | Line 157: | ||
What program do you use? | What program do you use? | ||
| - | 57°C auf 70°C ( | + | "57°C auf 70°C" (first annealing temperature: 58°C and then after 10 cycles 68°C) |
| Line 98: | Line 165: | ||
How did you label the PCR-Product, where is it stored and what do you do next? | How did you label the PCR-Product, where is it stored and what do you do next? | ||
| - | |||
| - | + | '''DNA-concentration measured with nanodrop:''' | |
| - | + | {| cellpadding="10" cellspacing="0" border="1" | |
| + | |sample | ||
| + | |DNA-concentration (ng/μl) | ||
| + | |- | ||
| + | |S35 | ||
| + | |75.1 | ||
| + | |- | ||
| + | |S45 | ||
| + | |45.1 | ||
| + | |} | ||
| + | ==<span style="color:grey;">Precipitator</span>== | ||
| - | + | Ruediger 27.07 | |
| + | picked colonies from yesterdays Trafo into medium with 2μl/ml Tet | ||
| - | + | S39-P18 | |
| - | + | S39-P19 | |
| - | + | S39-P20 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | S43-P18 | |
| + | S43-P19 | ||
| + | S43-P20 | ||
| - | + | overnight in 37°C | |
Latest revision as of 01:01, 22 September 2011
 "
"
 Contact
Contact