Team:WashU/Notebook/July2011
From 2011.igem.org
Bdonaldson (Talk | contribs) (→July 25) |
|||
| (62 intermediate revisions not shown) | |||
| Line 22: | Line 22: | ||
*Well 4 is positive control with Kan primers and Kan DNA | *Well 4 is positive control with Kan primers and Kan DNA | ||
*Well 5 just has PCR mix with the Nat primers. | *Well 5 just has PCR mix with the Nat primers. | ||
| + | |||
| + | Gel Team: | ||
| + | *Did a restriction digest on nat cassette using AgeI | ||
| + | *set up ligation reaction and let run over weekend | ||
== July 5 == | == July 5 == | ||
| Line 27: | Line 31: | ||
positive control contained mastermix, LEU2 primers and 100x diluted LEU2 DNA. | positive control contained mastermix, LEU2 primers and 100x diluted LEU2 DNA. | ||
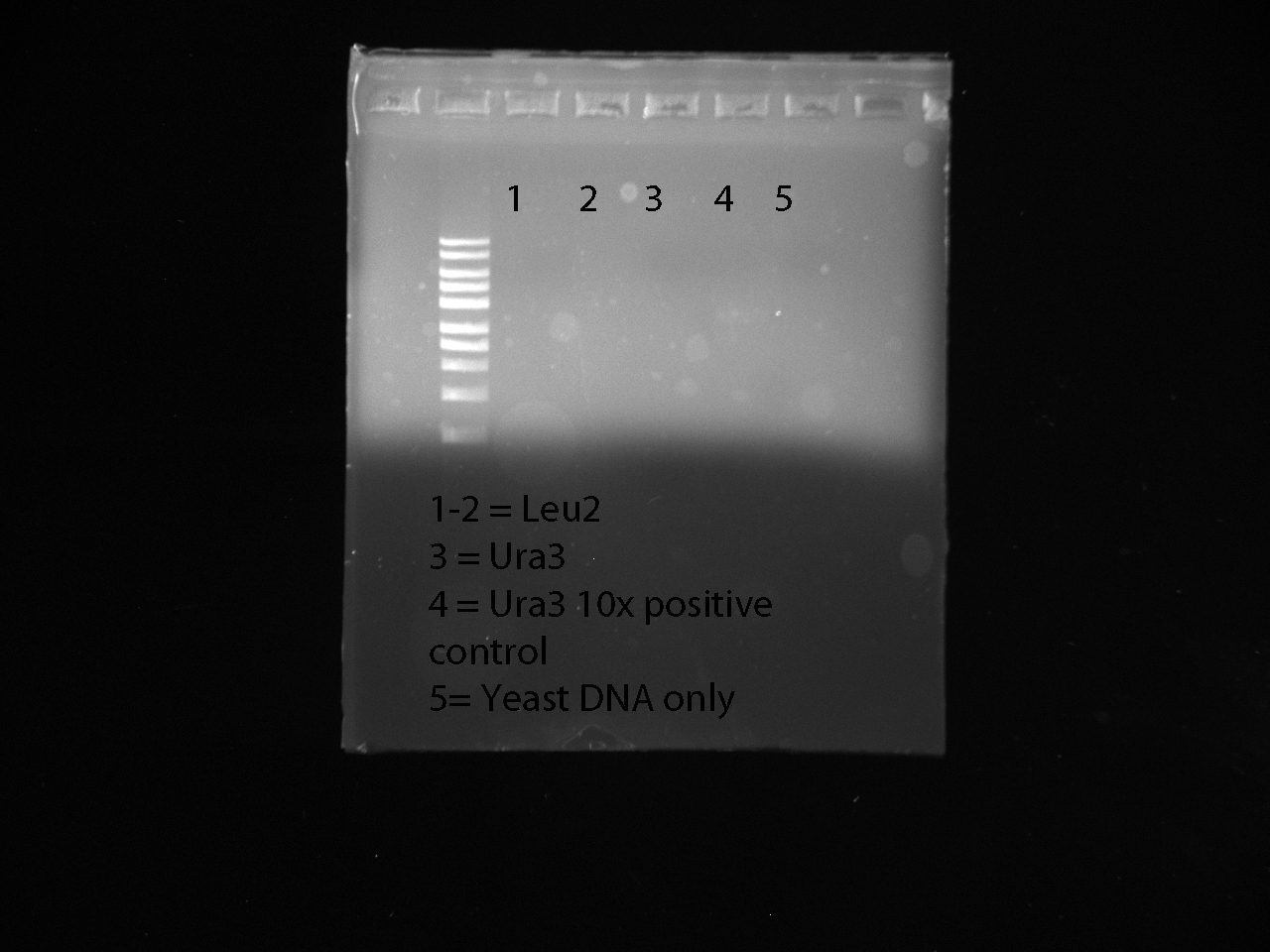
negative control contained mastermix, 100x diluted URA3 | negative control contained mastermix, 100x diluted URA3 | ||
| + | |||
| + | Gel Results: | ||
| + | |||
| + | [[File:CassettePCR 7-5 (Ura) copy.jpg|thumb|none|upright=3]] | ||
| + | |||
Note: this PCR reaction failed, including the positive control | Note: this PCR reaction failed, including the positive control | ||
| Line 39: | Line 48: | ||
**1.5 uL reverse primer | **1.5 uL reverse primer | ||
**1.0 ul diluted DNA | **1.0 ul diluted DNA | ||
| - | + | ||
| + | Gel Team | ||
| + | *ran a gel of the ligation reaction from 7/1 | ||
| + | **[https://3626779877665202800-a-1802744773732722657-s-sites.googlegroups.com/site/washuigem2011/files/5.jpg?attachauth=ANoY7crrAsrSGJwv64GdKkjnlUdwaJwjJ5ITE7xw6sSCy0hz7hG6LyjA86yasxYGizutyTwWiCUv5QJ6lqCcqW6gP51apVBUVQgthnd7WcJdCGVk8nIr5pORwUoG3xLK9g6jI8q1Ll5AzT3BokRYgLUvcplzJubLrYbktjQhbJgotzlKQQo31Tlgq3pfF6knRVKpiZlIsBvv&attredirects=0] | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Performed yeast growth curve on BC177 and BC178 to see how long it took for each strain to reach mid-log phase from a single colony on a YPD plate. | ||
| + | |||
| + | ==July 6== | ||
| + | Gel team: | ||
| + | *ran a gel of Leu 2 cassettes | ||
| + | **[https://3626779877665202800-a-1802744773732722657-s-sites.googlegroups.com/site/washuigem2011/files/6.jpg?attachauth=ANoY7cqCB9plxMPLpY-9Suw09cpHJxxcwNTfuw2ObI2NL9Z9ZzM7U3isUf5GDCNdxgR_P64EtKfKP_EJUK9kLoO_yYS4a2nAEMvsHCInCtR_4z87D6PrWrPMsIQYhtI8SB7ZsgURr-Fj7mcoRe1WXEg1_gNgy9FYDYYLhUPHVhf6P83ZKhR7gIEnk_4vidF5ApG_rACRbmB-&attredirects=0] | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Prepared samples of BC177 and BC178 from overnight cultures. Pelleted via centrifuging and lysed with glassed beads. The lysed yeast extract was analyzed in the spectrophotometer. | ||
| + | |||
== July 7 == | == July 7 == | ||
Note: the 10x dilution of URA3 was the most effective, positive with LEU2 failed. | Note: the 10x dilution of URA3 was the most effective, positive with LEU2 failed. | ||
| Line 51: | Line 77: | ||
1st negative control contained the mastermix and just LEU2 primers | 1st negative control contained the mastermix and just LEU2 primers | ||
2nd negative control negative contained the mastermix and just URA3 2x diluted DNA | 2nd negative control negative contained the mastermix and just URA3 2x diluted DNA | ||
| + | |||
| + | Gel Results: | ||
| + | |||
| + | [[File:CassettePCR 7-7 (Ura).jpg|thumb|none|upright=3]] | ||
PCR Protocol for Plasmids Review | PCR Protocol for Plasmids Review | ||
| Line 67: | Line 97: | ||
*In the meantime, we have designed an experiment to confirm that we have the pRS425 plasmid. We will cut the plasmid with specific restriction enzymes and run the digest on a gel to measure the resulting lengths. | *In the meantime, we have designed an experiment to confirm that we have the pRS425 plasmid. We will cut the plasmid with specific restriction enzymes and run the digest on a gel to measure the resulting lengths. | ||
**Of the restriction enzymes we have, we find that BamHI, XbaI, PstI, and XhoI cut the plasmid once, and EcoRI cuts the plasmid twice. We will use EcoRI and compare the lengths of the two fragments. We will also cut with BamHI and run that in a separate lane to confirm the overall plasmid length. | **Of the restriction enzymes we have, we find that BamHI, XbaI, PstI, and XhoI cut the plasmid once, and EcoRI cuts the plasmid twice. We will use EcoRI and compare the lengths of the two fragments. We will also cut with BamHI and run that in a separate lane to confirm the overall plasmid length. | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Prepared samples of beta-carotene stock solution in sterile water (8,16,32 microliter). We tried extracting beta-carotene from the water solution using hexane. Our results indicated very low yield from this extraction. This may be attributed to beta-carotene might have degraded due to prolonged exposure to light. | ||
== July 8 == | == July 8 == | ||
| Line 82: | Line 116: | ||
***Lane 6: 5 uL control | ***Lane 6: 5 uL control | ||
***Ran it for one hour at 132 volts | ***Ran it for one hour at 132 volts | ||
| + | Gel Team: | ||
| + | did a restriction digest of nat cassette with AgeI | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Prepared 3 5mL samples of lysed and saponified yeast cells from BC177 and BC178. | ||
| + | **Yeast samples came from overnight cultures. | ||
| + | **We followed extraction procedure from “High Level Production of Beta-Carotene in Saccharomyces cerevisiae…” Verwaal et. al. (2007). | ||
| + | **This procedure used transformed yeast, we used untransformed BC177 and BC178 cells. | ||
| + | **We did not add any beta-carotene. | ||
| + | *Our spectrophotometer analysis showed that the hexane layer contained minute amounts of lysed yeast cell product. These products did not interfere in the 453 nm range (where beta-carotene’s peak is). | ||
== July 11 == | == July 11 == | ||
| Line 100: | Line 145: | ||
**1.5 uL reverse primer | **1.5 uL reverse primer | ||
**1.0 ul diluted DNA | **1.0 ul diluted DNA | ||
| + | |||
| + | Gel Team: | ||
| + | the three empty wells on the right are our failed restriction digest | ||
| + | [[File:CassettePCR 7-11 (Ura).jpg|thumb|none|upright=3]] | ||
Transformation: | Transformation: | ||
*Set up yeast overnight for BC178 | *Set up yeast overnight for BC178 | ||
*Ran gel again with lower concentration enzyme; it did not work again | *Ran gel again with lower concentration enzyme; it did not work again | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Prepared 3 5mL samples of lysed and saponified yeast cells from BC177 and BC178. | ||
| + | **Yeast samples came from overnight cultures. We followed extraction procedure from “High Level Production of Beta-Carotene in Saccharomyces cerevisiae…” Verwaal et. al. (2007). | ||
| + | **This procedure used transformed yeast, we instead added known concentrations of beta-carotene (8,16,32 microliter for each strain). | ||
| + | *Our results indicated that the beta-carotene was not evenly distributed in the hexane layer. | ||
| + | *We modified the procedure so that we can get even distribution of beta-carotene in the hexane layer by pipetting the hexane layer up and down. | ||
| + | |||
*E. coli transformation of Leu2 and Ura3 and plated it with Leu2-deficient drop-out media we made | *E. coli transformation of Leu2 and Ura3 and plated it with Leu2-deficient drop-out media we made | ||
| Line 111: | Line 169: | ||
*Yeast transformation for BC178; we accidentally added 1 M LiAC instead of 100 uM, so we might have to redo it | *Yeast transformation for BC178; we accidentally added 1 M LiAC instead of 100 uM, so we might have to redo it | ||
*Set up E. coli overnight | *Set up E. coli overnight | ||
| + | |||
| + | Gel Team: | ||
| + | *did a restriction digest on nat cassette using AgeI | ||
| + | *ran digest on gel and excised the two bands for later gel extraction | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Repeated procedure from 7/11/11 procedure with modifications. | ||
| + | **Using hexane reagent as a blank. | ||
| + | *Much better results; absorbance almost doubled perfectly when beta-carotene concentrations were doubled. | ||
== July 13 == | == July 13 == | ||
| Line 141: | Line 209: | ||
We made 2 samples with Leu2 primers, 2 samples with Uras3 primers, a positive control with Uras3 primers and DNA, and a negative control with just yeast DNA. | We made 2 samples with Leu2 primers, 2 samples with Uras3 primers, a positive control with Uras3 primers and DNA, and a negative control with just yeast DNA. | ||
Strain 827 was used | Strain 827 was used | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Experimented with different blanks | ||
| + | **Prepared a blank by adding only yeast cells, lysing them, and adding hexane. We took an aliquot of the hexane layer from the cell lysate tube and used it as a blank. | ||
| + | **Absorbances for this procedure were much lower. | ||
| + | *Concluded that blanks have a small effect on the absorbance | ||
| + | *Performed this procedure on both strains of yeast | ||
| + | **However the blank was made from only one | ||
| + | **The tubes from which we got our two strains of yeast were grown to slightly different optical densities. This made one strain of yeast to have a higher concentration of yeast than the other. | ||
| + | **This may explain why we got negative absorbances in the 550-750nm range | ||
| + | *Same procedure was tried without using pyrogallol in methanol and KOH. This might have also contributed to the low absorbances | ||
| + | *We inferred from the data that pyrogallol and KOH were necessary reagents in the procedure | ||
== July 14 == | == July 14 == | ||
| Line 171: | Line 252: | ||
We made 1 samples with Leu2 primers, 2 samples with Uras3 primers and a positive control with Uras3 primers and DNA | We made 1 samples with Leu2 primers, 2 samples with Uras3 primers and a positive control with Uras3 primers and DNA | ||
Strain 825 was used | Strain 825 was used | ||
| + | |||
| + | Gel Team: | ||
| + | *ran a gel of the colony PCR | ||
| + | [[File:Colony PCR 7-14.jpg|thumb|none|upright=3]] | ||
| + | Gel Team: | ||
| + | *ran a gel of the colony PCR | ||
| + | **[[File:Colony PCR 7-13.gif|none|thumb|upright=3]] | ||
| + | *did a gel extraction with grad student Bert Berla | ||
| + | **[[File:sesamstrasse_bert.jpg|thumb|none|upright=2]] | ||
| + | *ran a ligation reaction overnight | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Tested for the effect of methanol in the procedure | ||
| + | **Extracted beta-carotene using previous procedure using pyrogallol dissolved in methanol and in water | ||
| + | **Used KOH and used hexane as a blank | ||
| + | *Concluded that methanol along with hexane helps extract the beta-carotene from the cell lysate products. In the samples with water instead of methanol, the spectra had much lower absorbances and had different shaped slopes in the 550-750nm range | ||
== July 15 == | == July 15 == | ||
Ordered Biobrick primers and all 4 cassette primers without homology in order to run colony PCR | Ordered Biobrick primers and all 4 cassette primers without homology in order to run colony PCR | ||
| + | |||
| + | Gel Team: | ||
| + | *ran a gel with PCR product from 7/13 and our ligation reaction from 7/14 | ||
| + | **[[https://3626779877665202800-a-1802744773732722657-s-sites.googlegroups.com/site/washuigem2011/files/14.jpg?attachauth=ANoY7cosx46T8ZSWGL2MZxuaA5kIny3T5og9EAmTXuyj7VV0bpDDrkyGJH2BLHeNXoY5ZBJ324GXSgelqV6Gnjg87LjFIqd3jtGjhbMvjGdukimu-Zvqq07S12zu8Mhz_e7XXlfmaNF13Ye9Kwrtq3Pi7hFUahe4khsHy7wnWRa-zF1jJ22JW5d9PMzYP1zUpm-OOqirtl2f&attredirects=0]] | ||
== July 18 == | == July 18 == | ||
| Line 187: | Line 289: | ||
*16.5 uL dH20 | *16.5 uL dH20 | ||
*0.25 uL Accutaq LA DNA polyermerase | *0.25 uL Accutaq LA DNA polyermerase | ||
| + | |||
| + | |||
| + | Gel Results: | ||
| + | |||
| + | [[File:Gene PCR (1st half) 7-18.jpg|thumb|none|upright=3]] | ||
| + | [[File:Gene PCR (2nd half) 7-18 copy.jpg|thumb|none|upright=3]] | ||
| + | |||
Transformation: | Transformation: | ||
| Line 205: | Line 314: | ||
*16.5 uL dH20 | *16.5 uL dH20 | ||
*0.25 uL Accutaq LA DNA polyermerase | *0.25 uL Accutaq LA DNA polyermerase | ||
| + | |||
| + | Gel Results: | ||
| + | |||
| + | [[File:CassettePCR 7-20 (ura-leu-kan-nat) copy.jpg|thumb|none|upright=3]] | ||
Designed primers to amplify Homology, Gene, Cassette, Homology | Designed primers to amplify Homology, Gene, Cassette, Homology | ||
| Line 247: | Line 360: | ||
*E. coli transformations using Nat and Kan | *E. coli transformations using Nat and Kan | ||
*Set up yeast overnights to try again. The contaminated plates must have yeast in them that could somehow grow in Leu-deficient medium. We are plating again to verify. | *Set up yeast overnights to try again. The contaminated plates must have yeast in them that could somehow grow in Leu-deficient medium. We are plating again to verify. | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Designed an experiment to specifically gauge the effects of different blanks. | ||
| + | **Used pure hexane reagent, mixture of hexane and methanol, and hexane layer from cell lysate. | ||
| + | *Tested these different blanks on 3 samples with the same concentration of beta-carotene. | ||
| + | *In the middle of the procedure, the cell lysate blank was spilled. We could not finish the experiment. | ||
| + | |||
| + | ==7/21/11== | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Re-did the experiment from yesterday on the blanks | ||
| + | *Using the results from this experiment we used the appropriate blank for another experiment to gauge our extraction efficiency | ||
| + | *Prepared four samples of each concentration of beta-carotene (2,4,8,16,32,64 µl of stock). | ||
| + | *Determined out extraction efficiency | ||
| + | |||
| + | Gel Team: | ||
| + | *ran a ligation reaction of nat fragments | ||
| + | *ran gel of ligation products: [https://mail.google.com/mail/u/0/?ui=2&ik=461b5e4071&view=att&th=1314e0f0d22ae774&attid=0.1&disp=inline&realattid=f_gqe31z8l0&zw ligation gel] | ||
| + | |||
| + | ==July 22== | ||
| + | |||
| + | PCR team: Realized the reverse primer design from July 20 was wrong. The redesigned primers are as follows: | ||
| + | |||
| + | **'''crtYB-URa3''' | ||
| + | *Reverse primer AGTATCATACTGTTCGTATACATACTTACTGACA | ||
| + | |||
| + | **BioBrick: | ||
| + | *Reverse biobrick: ctgcagcggccgcractagta AGTATCATACTGTTCGTATACATACTTACTGACA | ||
| + | |||
| + | **'''crtI-kanMX4''' | ||
| + | *Reverse primer ATGAACATATTCCATTTTGTAATTTCGTGTC | ||
| + | |||
| + | **Biobrick: | ||
| + | *Reverse biobrick primer: ctgcagcggccgcractagta ATGAACATATTCCATTTTGTAATTTCGTGTC | ||
| + | |||
| + | **'''CrtE-natMX4 ''' | ||
| + | *Reverse primer GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | ||
| + | |||
| + | **Biobrick: | ||
| + | *ctgcagcggccgcractagta GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT | ||
| + | |||
| + | **'''CCD1-leu2''' | ||
| + | *Forward primer AGTTACTCACTAATGACTAACGAAAAGGTCTG | ||
| + | *Reverse primer ATATTTAATTATTGTACATGGACATATCATACGTAA | ||
| + | |||
| + | **Biobricks: | ||
| + | *gaattcgcggccgcttctagag AGTTACTCACTAATGACTAACGAAAAGGTCTG | ||
| + | *ctgcagcggccgcractagta ATATTTAATTATTGTACATGGACATATCATACGTAA | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Redid calibration curve of beta-carotene in hexane | ||
| + | *Found that much of the beta-carotene has degraded | ||
| + | *Concluded that low efficiency in extraction is partly attributed to the deration | ||
| + | |||
| + | ==July 25== | ||
| + | |||
| + | PCR: | ||
| + | |||
| + | Nanodrop the minipreped Nat and Kan cassette | ||
| + | *Nat 1: 88.9 ng/uL | ||
| + | *Nat 2: 124.8 ng/uL | ||
| + | *Nat 3: 99.2 ng/uL | ||
| + | *Nat 4: 77.5 ng/uL | ||
| + | *Kan 1: 86.9 ng/uL | ||
| + | *Kan 2: 83.9 ng/uL | ||
| + | *Kan 3: 94.6 ng/uL | ||
| + | *Kan 4: 90.2 ng/uL | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Made a .1 mM beta-ionone stock solution in hexane through serial dilution | ||
| + | *Took spectrophotometric readings of samples of 1 ml of hexane with varying amounts (2, 4, 8, 16, 32, 64, 128, 256, 512 µl) of stock solution to make a calibration curve for beta-ionone in hexane | ||
| + | |||
| + | ==July 26== | ||
| + | |||
| + | Gel Team: | ||
| + | *ran a ligation of nat fragments | ||
| + | *ran ligation product on gel: [https://mail.google.com/mail/u/0/?ui=2&ik=461b5e4071&view=att&th=131675217293ad05&attid=0.1&disp=inline&realattid=f_gql3dz510&zw ligation] | ||
| + | **although not clear from image, ligation was success | ||
| + | |||
| + | ==July 27== | ||
| + | Transformation: | ||
| + | *We tried to determine the source of contamination on our plates. We plated our six transformation reagents on a YPD, Leu-dropout plate; plated ddH20, 100 mM LiAc, 1 M LiAc, DNA, PEG, YPD. | ||
| + | *Also, to prevent future contamination, we autoclaved some flasks and test tubes. | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Repeated yesterday’s procedure, except using a newly made bottle of beta-ionone stock, as beta-ionone readily degrades in light and air | ||
| + | |||
| + | ==July 28== | ||
| + | Transformation: | ||
| + | *There was yeast growing on our 100 mM LiAc streak. We made a new stock by diluting 4 mL 1 M LiAc with 36 mL ddH20 and filter-sterilized it. | ||
| + | *Still waiting on salmon-sperm DNA. | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Tested the extraction efficiency beta ionone in hexane from yeast extract by spectrophotometer. | ||
| + | **32µl of 0.1mM beta ionone solution was added to yeast cells suspended in 1ml of water and subsequently lysed with glass beads and extracted with 2ml of hexane | ||
| + | ** Measured absorbance on spectrophotometer and used absorbance at λmax (280nm) along with extinction coefficient obtained from calibration curve made on 7/26/11 | ||
| + | |||
| + | ==7/29/11== | ||
| + | |||
| + | Assay Preparation: | ||
| + | |||
| + | *Tested the limit of detection for beta ionone in yeast using varying amounts of beta ionone stock solution (2, 4, 8 µl) and following procedure from 7/28 | ||
| + | *Unfortunately, the spectrophotometer was unavailable. | ||
| + | *Samples were parafilmed and put in -20C freezer for weekend storage | ||
Latest revision as of 04:45, 28 September 2011
July 1
Master mix composition (6 wells worth)
- 15.0uL 10x buffer
- 7.5 uL dNTP
- 99 uL dH20
- 1.5 uL Accutaq LA DNA polyermerase
= 20.5 ul/well
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 10ng Kan DNA (1mL using our 2x diluted sample)
- 1.5ul of Nat ligation product
2nd attempt at PCR on the Nat ligation product:
- Nat was not detected on a gel but may be because a very small amount of product was made.
- Well 1,2,3 are PCR mix with Nat primers and 1.5uL of Nat ligation product per well
- Well 4 is positive control with Kan primers and Kan DNA
- Well 5 just has PCR mix with the Nat primers.
Gel Team:
- Did a restriction digest on nat cassette using AgeI
- set up ligation reaction and let run over weekend
July 5
Ran PCR to amplify 100x diluted sample of URA3 positive control contained mastermix, LEU2 primers and 100x diluted LEU2 DNA. negative control contained mastermix, 100x diluted URA3
Gel Results:
Note: this PCR reaction failed, including the positive control
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
Gel Team
- ran a gel of the ligation reaction from 7/1
Assay Preparation:
- Performed yeast growth curve on BC177 and BC178 to see how long it took for each strain to reach mid-log phase from a single colony on a YPD plate.
July 6
Gel team:
- ran a gel of Leu 2 cassettes
Assay Preparation:
- Prepared samples of BC177 and BC178 from overnight cultures. Pelleted via centrifuging and lysed with glassed beads. The lysed yeast extract was analyzed in the spectrophotometer.
July 7
Note: the 10x dilution of URA3 was the most effective, positive with LEU2 failed. Reran PCR reaction to amplify different concentrations of URA3. For this reaction we used a 2x, 5x, 10x, and 15x diluted samples of URA3 - ran each sample twice 2x: 1.5ul DNA, 1.5ul dH20 5x: 1ul DNA, 4ul dH20 10x: 1ul DNA, 9ul dH20 15x: 1ul DNA, 14ul dH20 positive control contained mastermix, LEU2 primers, and 20x diluted LEU2 DNA 1st negative control contained the mastermix and just LEU2 primers 2nd negative control negative contained the mastermix and just URA3 2x diluted DNA
Gel Results:
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
Transformation:
- We are still waiting for Leu2 dropout medium to come in.
- In the meantime, we have designed an experiment to confirm that we have the pRS425 plasmid. We will cut the plasmid with specific restriction enzymes and run the digest on a gel to measure the resulting lengths.
- Of the restriction enzymes we have, we find that BamHI, XbaI, PstI, and XhoI cut the plasmid once, and EcoRI cuts the plasmid twice. We will use EcoRI and compare the lengths of the two fragments. We will also cut with BamHI and run that in a separate lane to confirm the overall plasmid length.
Assay Preparation:
- Prepared samples of beta-carotene stock solution in sterile water (8,16,32 microliter). We tried extracting beta-carotene from the water solution using hexane. Our results indicated very low yield from this extraction. This may be attributed to beta-carotene might have degraded due to prolonged exposure to light.
July 8
Transformation:
- We digested the DNA and then ran the results on a gel.
- Into a dilution microcentrifuge tube, we used 2 uL DNA (concentration 235.4 ng/uL) and 248 uL ddH20.
- BamHI tube: 2 uL diluted DNA, 2 uL NEBuffer, 2 uL diluted BSA (10x), 1 uL BamHI enzyme, 13 uL ddH20
- EcoRI tube: 2 uL diluted DNA, 2 ul SH buffer, 1 uL EcoRI enzyme, 15 uL reaction
- Incubated both tubes in a 37 C water bath for one hour for digestion
- Ran the solutions on a gel
- 5 uL loading dye into three tubes: BamHI, EcoRI, control (uncut plasmid DNA)
- Lane 1: 5 uL ladder
- Lanes 2 and 3: 5 uL each BamHI
- Lanes 4 and 5: 5 uL each EcoRI
- Lane 6: 5 uL control
- Ran it for one hour at 132 volts
Gel Team: did a restriction digest of nat cassette with AgeI
Assay Preparation:
- Prepared 3 5mL samples of lysed and saponified yeast cells from BC177 and BC178.
- Yeast samples came from overnight cultures.
- We followed extraction procedure from “High Level Production of Beta-Carotene in Saccharomyces cerevisiae…” Verwaal et. al. (2007).
- This procedure used transformed yeast, we used untransformed BC177 and BC178 cells.
- We did not add any beta-carotene.
- Our spectrophotometer analysis showed that the hexane layer contained minute amounts of lysed yeast cell product. These products did not interfere in the 453 nm range (where beta-carotene’s peak is).
July 11
PCR group: Reran five wells of PCR on URA3 - used 10x dilution URA3 DNA in our wells. Positive control contained 2ul of 20x diluted LEU2, LEU2 primers, and mastermix. Negative control contained 1ul 10x dilution URA3 and mastermix --> check for Plasmid DNA
PCR Protocol for Plasmids Review
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
- 20.5 ul/well
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 1.0 ul diluted DNA
Gel Team: the three empty wells on the right are our failed restriction digest
Transformation:
- Set up yeast overnight for BC178
- Ran gel again with lower concentration enzyme; it did not work again
Assay Preparation:
- Prepared 3 5mL samples of lysed and saponified yeast cells from BC177 and BC178.
- Yeast samples came from overnight cultures. We followed extraction procedure from “High Level Production of Beta-Carotene in Saccharomyces cerevisiae…” Verwaal et. al. (2007).
- This procedure used transformed yeast, we instead added known concentrations of beta-carotene (8,16,32 microliter for each strain).
- Our results indicated that the beta-carotene was not evenly distributed in the hexane layer.
- We modified the procedure so that we can get even distribution of beta-carotene in the hexane layer by pipetting the hexane layer up and down.
- E. coli transformation of Leu2 and Ura3 and plated it with Leu2-deficient drop-out media we made
July 12
Transformation:
- Plates from yesterday
- Yeast transformation for BC178; we accidentally added 1 M LiAC instead of 100 uM, so we might have to redo it
- Set up E. coli overnight
Gel Team:
- did a restriction digest on nat cassette using AgeI
- ran digest on gel and excised the two bands for later gel extraction
Assay Preparation:
- Repeated procedure from 7/11/11 procedure with modifications.
- Using hexane reagent as a blank.
- Much better results; absorbance almost doubled perfectly when beta-carotene concentrations were doubled.
July 13
PCR:
Colony PCR:
Protocol:
- Transfer a yeast colony to a solution of 0.2% SDS. The SDS page was made by adding 0.02g SDS to 10mL of water.
- Vortex for 15 sec
- Heat in PCR machine for 4 min at 90 degrees Celsius.2
- Microcentrifuge for 1 min.
- Pipet out the supernatent and store at -20 degrees Celsius.
PCR mix for 50uL reaction:
- 5uL 10x PCR buffer
- 1.5 uL 50mM MgCl2 (we used 3uL because we 25mM MgCl2)
- 1uL of 10mM dNTPs
- 2uL of 25% TritonX-100
- 0.3 uL Taq polymersase
- 31.7 uL water
- Add 43 uL of PCR mix to each PCR tube
- Then add 3 uL of both the forward and reverse primers
- Add 1uL of the DNA from yeast
We made 2 samples with Leu2 primers, 2 samples with Uras3 primers, a positive control with Uras3 primers and DNA, and a negative control with just yeast DNA.
Strain 827 was used
Assay Preparation:
- Experimented with different blanks
- Prepared a blank by adding only yeast cells, lysing them, and adding hexane. We took an aliquot of the hexane layer from the cell lysate tube and used it as a blank.
- Absorbances for this procedure were much lower.
- Concluded that blanks have a small effect on the absorbance
- Performed this procedure on both strains of yeast
- However the blank was made from only one
- The tubes from which we got our two strains of yeast were grown to slightly different optical densities. This made one strain of yeast to have a higher concentration of yeast than the other.
- This may explain why we got negative absorbances in the 550-750nm range
- Same procedure was tried without using pyrogallol in methanol and KOH. This might have also contributed to the low absorbances
- We inferred from the data that pyrogallol and KOH were necessary reagents in the procedure
July 14
PCR:
Colony PCR:
Protocol:
- Transfer a yeast colony to a solution of 0.2% SDS. The SDS page was made by adding 0.02g SDS to 10mL of water.
- Vortex for 15 sec
- Heat in PCR machine for 4 min at 90 degrees Celsius.2
- Microcentrifuge for 1 min.
- Pipet out the supernatent and store at -20 degrees Celsius.
PCR mix for 50uL reaction:
- 5uL 10x PCR buffer
- 1.5 uL 50mM MgCl2 (we used 3uL because we 25mM MgCl2)
- 1uL of 10mM dNTPs
- 2uL of 25% TritonX-100. The 25% TritonX-100 was made by adding 6ul of TritonX-100 to 18ul of H20
- 0.3 uL Taq polymersase
- 31.7 uL water
- Add 43 uL of PCR mix to each PCR tube
- Then add 3 uL of both the forward and reverse primers
- Add 1uL of the DNA from yeast
We made 1 samples with Leu2 primers, 2 samples with Uras3 primers and a positive control with Uras3 primers and DNA
Strain 825 was used
Gel Team:
- ran a gel of the colony PCR
Gel Team:
- ran a gel of the colony PCR
- did a gel extraction with grad student Bert Berla
- ran a ligation reaction overnight
Assay Preparation:
- Tested for the effect of methanol in the procedure
- Extracted beta-carotene using previous procedure using pyrogallol dissolved in methanol and in water
- Used KOH and used hexane as a blank
- Concluded that methanol along with hexane helps extract the beta-carotene from the cell lysate products. In the samples with water instead of methanol, the spectra had much lower absorbances and had different shaped slopes in the 550-750nm range
July 15
Ordered Biobrick primers and all 4 cassette primers without homology in order to run colony PCR
Gel Team:
- ran a gel with PCR product from 7/13 and our ligation reaction from 7/14
- [[3]]
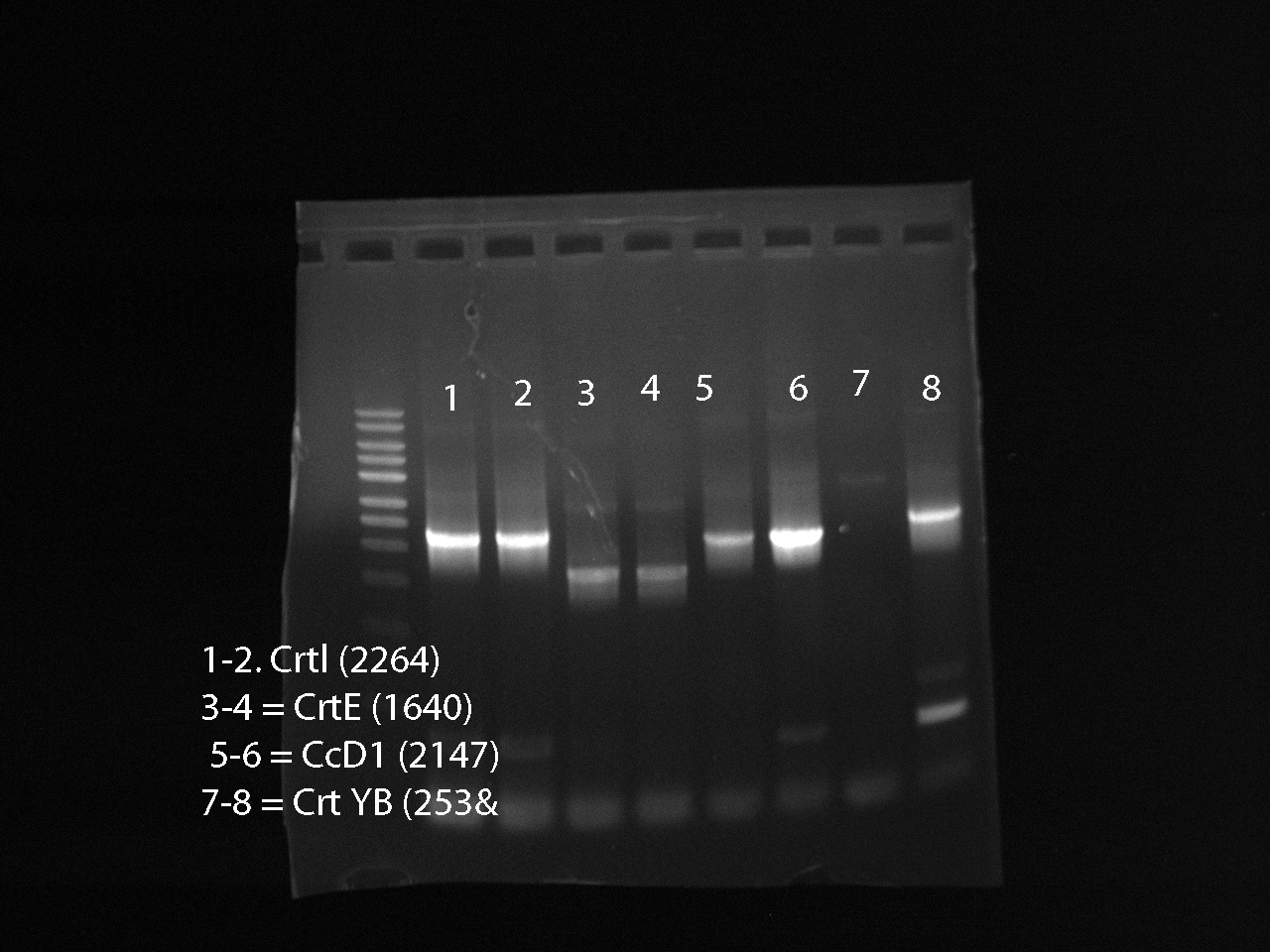
July 18
Ran a PCR for the genes: CrtI, CrtE, CcD1, CcYB --> ran 3 PCR tubes for each gene
- 1 positive control: Nat Genes + primers
- 4 negative controls, 1 corresponding to each gene, contains only the mastermix soln and F/R primers
Recipe per sample for running plasmid DNA:
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
Gel Results:
Transformation:
- We had made colonies of BC178, but we had gotten contamination in them, so we made new plates, this time with ampicillin, in hopes of getting rid of any E. coli that may have been in our test tubes. Also, we filter-sterilized our lithium acetate (just in case) and our PEG 3350.
- We set up new overnights to start over.
July 20
PCR Team: Ran a PCR for the cassettes: URA3, LEU2, KAN, NAT --> ran 2 41ul PCR tubes for each gene
- 4 25ul negative controls, 1 corresponding to each gene, contains only the mastermix soln and F/R primers
Recipe per sample for running plasmid DNA (20.5ul reactions):
- 2.5uL 10x buffer
- 1.25 uL dNTP
- 1.5 uL forward primer
- 1.5 uL reverse primer
- 16.5 uL dH20
- 0.25 uL Accutaq LA DNA polyermerase
Gel Results:
Designed primers to amplify Homology, Gene, Cassette, Homology
- crtYB-URa3
- Forward primer GAAGAATATACTAAAAAATGAGCAGGCAAGATA 33 54.78
- Reverse primer TATGAATGTCAGTAAGTATGTATACGAACAGTAT 34 54.58
- BioBrick:
- gaattcgcggccgcttctagag GAA GAA TAT ACT AAA AAA TGA GCA GGC AAG ATA
- ctgcagcggccgcractagta T ATG AAT GTC AGT AAG TAT GTA TAC GAA CAG TAT
- crtI-kanMX4
- Forward primer TTTTCCAATAGGTGGTTAGCAATCGTC 27 55.97
- Reverse primer AAATTCATAATAGAAACGACACGAAATTACAAA 33 54.14
- Biobrick:
- Gaattcgcggccgcttctagag TTT TCC AAT AGG TGG TTA GCA ATC GTC TTA
- Ctgcagcggccgcractagta AAA TTC ATA ATA GAA ACG ACA CGA AAT TAC AAA A
- CrtE-natMX4
- Forward primer TCTCCGAGCAGAAGGAAGAAC 21 53.62
- Reverse primer CTAAACTCACAAATTAGAGCTTCAATTTAATTATAT 36 52.82
- Biobricks:
- gaattcgcggccgcttctagag TCT CCG AGC AGA AGG AAG AAC
- ctgcagcggccgcractagta CTA AAC TCA CAA ATT AGA GCT TCA ATT TAA TTA TAT
- CCD1-leu2
- Forward primer AGTTACTCACTAATGACTAACGAAAAGGTCTG 32 56.97
- Reverse primer GGTTGAGCATTACGTATGATATGTCCATGT 30 57.38
- Biobricks:
- gaattcgcggccgcttctagag AGTTACTCACTAATGACTAACGAAAAGGTCTG
- ctgcagcggccgcractagta GGT TGA GCA TTA CGT ATG ATA TGT CCA TGT
Transformation:
- Made more YPD
- E. coli transformations using Nat and Kan
- Set up yeast overnights to try again. The contaminated plates must have yeast in them that could somehow grow in Leu-deficient medium. We are plating again to verify.
Assay Preparation:
- Designed an experiment to specifically gauge the effects of different blanks.
- Used pure hexane reagent, mixture of hexane and methanol, and hexane layer from cell lysate.
- Tested these different blanks on 3 samples with the same concentration of beta-carotene.
- In the middle of the procedure, the cell lysate blank was spilled. We could not finish the experiment.
7/21/11
Assay Preparation:
- Re-did the experiment from yesterday on the blanks
- Using the results from this experiment we used the appropriate blank for another experiment to gauge our extraction efficiency
- Prepared four samples of each concentration of beta-carotene (2,4,8,16,32,64 µl of stock).
- Determined out extraction efficiency
Gel Team:
- ran a ligation reaction of nat fragments
- ran gel of ligation products: ligation gel
July 22
PCR team: Realized the reverse primer design from July 20 was wrong. The redesigned primers are as follows:
- crtYB-URa3
- Reverse primer AGTATCATACTGTTCGTATACATACTTACTGACA
- BioBrick:
- Reverse biobrick: ctgcagcggccgcractagta AGTATCATACTGTTCGTATACATACTTACTGACA
- crtI-kanMX4
- Reverse primer ATGAACATATTCCATTTTGTAATTTCGTGTC
- Biobrick:
- Reverse biobrick primer: ctgcagcggccgcractagta ATGAACATATTCCATTTTGTAATTTCGTGTC
- CrtE-natMX4
- Reverse primer GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT
- Biobrick:
- ctgcagcggccgcractagta GGGTAATAACTGATATAATTAAATTGAAGCTCTAAT
- CCD1-leu2
- Forward primer AGTTACTCACTAATGACTAACGAAAAGGTCTG
- Reverse primer ATATTTAATTATTGTACATGGACATATCATACGTAA
- Biobricks:
- gaattcgcggccgcttctagag AGTTACTCACTAATGACTAACGAAAAGGTCTG
- ctgcagcggccgcractagta ATATTTAATTATTGTACATGGACATATCATACGTAA
Assay Preparation:
- Redid calibration curve of beta-carotene in hexane
- Found that much of the beta-carotene has degraded
- Concluded that low efficiency in extraction is partly attributed to the deration
July 25
PCR:
Nanodrop the minipreped Nat and Kan cassette
- Nat 1: 88.9 ng/uL
- Nat 2: 124.8 ng/uL
- Nat 3: 99.2 ng/uL
- Nat 4: 77.5 ng/uL
- Kan 1: 86.9 ng/uL
- Kan 2: 83.9 ng/uL
- Kan 3: 94.6 ng/uL
- Kan 4: 90.2 ng/uL
Assay Preparation:
- Made a .1 mM beta-ionone stock solution in hexane through serial dilution
- Took spectrophotometric readings of samples of 1 ml of hexane with varying amounts (2, 4, 8, 16, 32, 64, 128, 256, 512 µl) of stock solution to make a calibration curve for beta-ionone in hexane
July 26
Gel Team:
- ran a ligation of nat fragments
- ran ligation product on gel: ligation
- although not clear from image, ligation was success
July 27
Transformation:
- We tried to determine the source of contamination on our plates. We plated our six transformation reagents on a YPD, Leu-dropout plate; plated ddH20, 100 mM LiAc, 1 M LiAc, DNA, PEG, YPD.
- Also, to prevent future contamination, we autoclaved some flasks and test tubes.
Assay Preparation:
- Repeated yesterday’s procedure, except using a newly made bottle of beta-ionone stock, as beta-ionone readily degrades in light and air
July 28
Transformation:
- There was yeast growing on our 100 mM LiAc streak. We made a new stock by diluting 4 mL 1 M LiAc with 36 mL ddH20 and filter-sterilized it.
- Still waiting on salmon-sperm DNA.
Assay Preparation:
- Tested the extraction efficiency beta ionone in hexane from yeast extract by spectrophotometer.
- 32µl of 0.1mM beta ionone solution was added to yeast cells suspended in 1ml of water and subsequently lysed with glass beads and extracted with 2ml of hexane
- Measured absorbance on spectrophotometer and used absorbance at λmax (280nm) along with extinction coefficient obtained from calibration curve made on 7/26/11
7/29/11
Assay Preparation:
- Tested the limit of detection for beta ionone in yeast using varying amounts of beta ionone stock solution (2, 4, 8 µl) and following procedure from 7/28
- Unfortunately, the spectrophotometer was unavailable.
- Samples were parafilmed and put in -20C freezer for weekend storage
 "
"