Team:Cambridge/Experiments/Initial Exercise Group Alpha
From 2011.igem.org
(→Fusion of SrfA promoter with GFP) |
|||
| (15 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
===Aim=== | ===Aim=== | ||
| - | Our week 1 experimental challenge was to generate a temporal and or spatial pattern of GFP expression in ''Bacillus subtilis'', using Gibson Assembly to join a sequence of interest to a GFP coding plasmid which we will amplify in E. coli. We chose to visualise sporulation via a fusion of SrfA promoter with GFP. | + | Our week 1 experimental challenge was to generate a temporal and/or spatial pattern of GFP expression in ''Bacillus subtilis'', using Gibson Assembly to join a sequence of interest to a GFP coding plasmid which we will amplify in E. coli. We chose to visualise sporulation-dependent expression via a fusion of SrfA promoter with GFP. |
===Role of SrfA=== | ===Role of SrfA=== | ||
| Line 19: | Line 19: | ||
===Predicted Results=== | ===Predicted Results=== | ||
| - | *In E. coli we do not expect to see any fluorescence. | + | *In E. coli we do not expect to see any fluorescence, though there is a slight chance that the B. Subtilis promoter we're using will be active to some extent in this species. |
*Bacillus colonies exhibit a range of colony patterns and superimposed on the colony shape we expect a temporal pattern of green fluorescence marking sporulation activity. | *Bacillus colonies exhibit a range of colony patterns and superimposed on the colony shape we expect a temporal pattern of green fluorescence marking sporulation activity. | ||
*intially zero or low GFP detection, higher GFP activity on activation of SrfA prior to sporulation. | *intially zero or low GFP detection, higher GFP activity on activation of SrfA prior to sporulation. | ||
| - | * | + | *Fluorescence will mark spatially sporulation sites. |
===Materials=== | ===Materials=== | ||
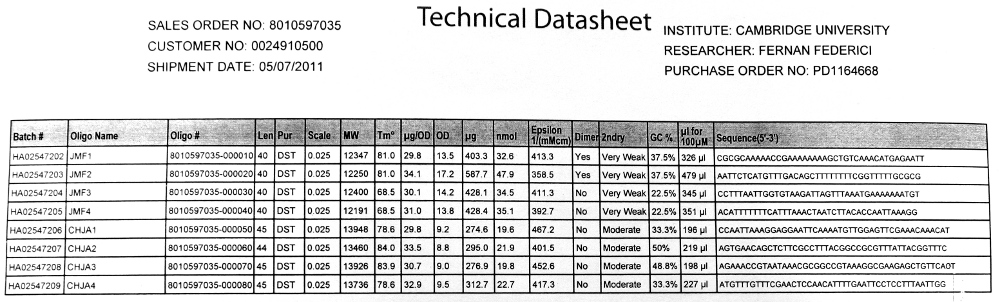
| - | [[File:CAM HJCA PRIMERS.JPG|thumb|400px|right| | + | [[File:CAM HJCA PRIMERS.JPG|thumb|400px|right|Primer datasheet]] |
*20bp primers with 20bp tails were designed to perform [[Team:Cambridge/Protocols/PCR | PCR]] to amplify our chosen DNA sequences and [[Team:Cambridge/Protocols/Gibson_Assembly | Gibson Assembly]] to fuse the SrfA promoter with a plasmid containing GFP. | *20bp primers with 20bp tails were designed to perform [[Team:Cambridge/Protocols/PCR | PCR]] to amplify our chosen DNA sequences and [[Team:Cambridge/Protocols/Gibson_Assembly | Gibson Assembly]] to fuse the SrfA promoter with a plasmid containing GFP. | ||
| - | We used the Finnzymes melting temperature calculator to work out the melting temperature of the primer part (not the tail) of our Gibson Assembly oligos (this was the 3' 20bp of each oligo) these values in ºC are shown below | + | We used the 'Finnzymes' melting temperature calculator to work out the melting temperature of the primer part (not the tail) of our Gibson Assembly oligos (this was the 3' 20bp of each oligo) these values in ºC are shown below |
JMF1 - 56.91 | JMF1 - 56.91 | ||
| Line 66: | Line 66: | ||
for a total volume 25μl | for a total volume 25μl | ||
| - | [[File:Exp1_panzer_results.jpg|200px|thumb|left| | + | [[File:Exp1_panzer_results.jpg|200px|thumb|left|Results from PCR reaction using taq]] |
The complete tubes were run in a real-time PCR machine for 30 cycles with a 2 minute extension time and a primer annealing temperature of 50ºc. Two of these were successfully copied but one of the plasmid fragments failed. The failure of this largest fragment to amplify was attributed to a too short extension time. | The complete tubes were run in a real-time PCR machine for 30 cycles with a 2 minute extension time and a primer annealing temperature of 50ºc. Two of these were successfully copied but one of the plasmid fragments failed. The failure of this largest fragment to amplify was attributed to a too short extension time. | ||
| Line 73: | Line 73: | ||
We used the same primers and templates as above, but we made a new master mix including phusion, a more processive and higher fidelity polymerase. Each PCR tube contained a total volume 25μl, which was comprised of: | We used the same primers and templates as above, but we made a new master mix including phusion, a more processive and higher fidelity polymerase. Each PCR tube contained a total volume 25μl, which was comprised of: | ||
| - | + | *5μl buffer | |
| - | 0. | + | *0.5μl dNTP |
| - | 0. | + | *0.25μl phusion polymerase |
| - | 16. | + | *16.25μl H2O |
| - | 1μl of each primer | + | *1μl of each primer |
| - | 1μl of template DNA | + | *1μl of template DNA |
We used gel electrophoresis to check the results and isolate our fragments. | We used gel electrophoresis to check the results and isolate our fragments. | ||
| - | |||
===Gibson Assembly=== | ===Gibson Assembly=== | ||
| Line 112: | Line 111: | ||
Following Gibson Assembly we transformed E.coli with the DNA we had combined. See the protocols page for how to do [[Team:Cambridge/Protocols/Transformation_of_E.Coli| Transformation of E.coli]]. We transformed and plated our E. coli on a medium containing ampicillin and left them to incubate overnight. In the morning we observed 5 colonies on our plate. | Following Gibson Assembly we transformed E.coli with the DNA we had combined. See the protocols page for how to do [[Team:Cambridge/Protocols/Transformation_of_E.Coli| Transformation of E.coli]]. We transformed and plated our E. coli on a medium containing ampicillin and left them to incubate overnight. In the morning we observed 5 colonies on our plate. | ||
| - | To our surprise, the E. coli colonies did exhibit fluorescence when viewed under a light microscope. | + | To our surprise, the E. coli colonies did exhibit fluorescence when viewed under a light microscope - the B. Subtilis promoter has cross-species activity. |
| + | |||
| + | [[File:cam_JMFexperiment_greencolonies.jpg | left | thumb | 380px | ''E.coli'' colonies]] | ||
| + | [[File:cam_JMFexperiment_greencolony.jpg | right | thumb | 380px | A single ''E.coli'' colony]] | ||
| + | |||
| + | <br style="clear:both;"/> | ||
===Digestion with Restriction Enzymes=== | ===Digestion with Restriction Enzymes=== | ||
| Line 128: | Line 132: | ||
The predicted restriction maps, prepared using ApE editor, are presented below. | The predicted restriction maps, prepared using ApE editor, are presented below. | ||
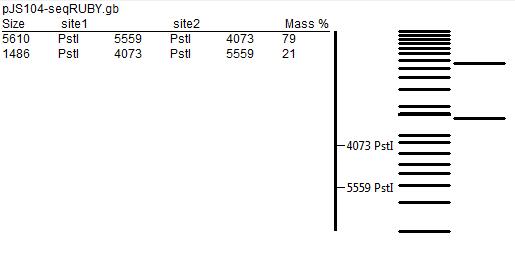
| - | [[File:cam_restrictionmap_JMFexperiment.jpg | centre | thumb | 400px | | + | [[File:cam_restrictionmap_JMFexperiment.jpg | centre | thumb | 400px | Restriction map of the plasmid with SrfA promoter:GFP fusion]] |
| - | [[File:cam_restrictionmap_positivecontrol.jpg | centre | thumb | 400px | | + | [[File:cam_restrictionmap_positivecontrol.jpg | centre | thumb | 400px | Restriction map of the positive control plasmid with mRUBY sequence]] |
| - | [[File:cam_restrictionmap_pJS104plasmid.jpg | centre | thumb | 400px | | + | [[File:cam_restrictionmap_pJS104plasmid.jpg | centre | thumb | 400px | Restriction map of the original pJS104 plasmid]] |
| - | We separated products of digestion by [[Team:Cambridge/Protocols/Gel_Electrophoresis | gel electrophoresis]] (using 1% agarose gel). As we can see in the picture on the left, we did not obtain clear bands. This was most probably caused by poor quality of the gel or unfavourable concentration of the buffer. Additionally, the number of smeared bands and their distribution is not consistent with predictions from ApE, and on some lanes with uncut vectors (e.g. lane 14)we can see several bands of different sizes, although a single band was expected. | + | We separated products of digestion by [[Team:Cambridge/Protocols/Gel_Electrophoresis | gel electrophoresis]] (using 1% agarose gel). As we can see in the picture on the left, we did not obtain clear bands. This was most probably caused by poor quality of the gel or unfavourable concentration of the buffer. Additionally, the number of smeared bands and their distribution is not consistent with predictions from ApE, and on some lanes with uncut vectors (e.g. lane 14) we can see several bands of different sizes, although a single band was expected. |
[[File:cam_firstgel_restrictiondigestion.jpg | centre | thunb | 500px | picture of an agarose gel with products of restriction enzyme digestion]] | [[File:cam_firstgel_restrictiondigestion.jpg | centre | thunb | 500px | picture of an agarose gel with products of restriction enzyme digestion]] | ||
| Line 139: | Line 143: | ||
A table describing the content of each lane (numbered from the left to the right): | A table describing the content of each lane (numbered from the left to the right): | ||
| - | {|border="1px" | + | {|border="1px" align="center" style="text-align:center;" |
!Lane | !Lane | ||
!1 | !1 | ||
Latest revision as of 13:59, 21 September 2011
Contents |
Fusion of SrfA promoter with GFP
Aim
Our week 1 experimental challenge was to generate a temporal and/or spatial pattern of GFP expression in Bacillus subtilis, using Gibson Assembly to join a sequence of interest to a GFP coding plasmid which we will amplify in E. coli. We chose to visualise sporulation-dependent expression via a fusion of SrfA promoter with GFP.
Role of SrfA
- SrfA codes for surfactin production, a protein that is produced by B. subtilis prior to sporulation.
- SrfA is regulated by ComA, a protein which itself responds to an intercellular population-density-signalling molecule, CSF, in a concentration dependent manner.
- ComA has low activity at low CSF concentration(low population density), highly active as CSF levels increase, and less active at high CSF concentrations.
Construct Design
We wish to:
- keep the original promotor for SrfA and comA boxes
- remove GFP promotor sequence
- maintain ComGA stabiliser sequence
- do not include restriction sites
Predicted Results
- In E. coli we do not expect to see any fluorescence, though there is a slight chance that the B. Subtilis promoter we're using will be active to some extent in this species.
- Bacillus colonies exhibit a range of colony patterns and superimposed on the colony shape we expect a temporal pattern of green fluorescence marking sporulation activity.
- intially zero or low GFP detection, higher GFP activity on activation of SrfA prior to sporulation.
- Fluorescence will mark spatially sporulation sites.
Materials
- 20bp primers with 20bp tails were designed to perform PCR to amplify our chosen DNA sequences and Gibson Assembly to fuse the SrfA promoter with a plasmid containing GFP.
We used the 'Finnzymes' melting temperature calculator to work out the melting temperature of the primer part (not the tail) of our Gibson Assembly oligos (this was the 3' 20bp of each oligo) these values in ºC are shown below
JMF1 - 56.91 JMF2 - 68.8 JMF3 - 50.48 JMF4 - 53.09
- 100 μM stock solutions of the primers were made and subsequently diluted four-fold to make 25μM 'working solutions'
- Particular care was taken in preparing the initial stock as these affect all future dilutions and were 'contingency' solutions in case of mistake.
- Genomic DNA of B. subtilis were provided and the vector plasmid pJS104-sfGFP containing the superfolded GFP gene.
- The two primers for two fragments of the plasmid were provided.
PCR -- taq
Our three PCR reactions consisted of:
- 1.
- 1μl primer JMF2
- 1μl primer JMF3
- 1μl B. subtilis genomic DNA
- 2.
- 1μl primer JMF1
- 1μl primer A Forward (provided)
- 1μl vector DNA
- 3
- 1μl primer JMF4
- 1μl primer B reverse (provided)
- 1μl vector DNA
to each tube we also added:
- 9.5μl of water
- 12.5μl Master mix (SyBR Green and Rox, Hotstart Taq polymerase, dNTPs and dyes)
for a total volume 25μl
The complete tubes were run in a real-time PCR machine for 30 cycles with a 2 minute extension time and a primer annealing temperature of 50ºc. Two of these were successfully copied but one of the plasmid fragments failed. The failure of this largest fragment to amplify was attributed to a too short extension time.
PCR -- Phusion(TM)
We used the same primers and templates as above, but we made a new master mix including phusion, a more processive and higher fidelity polymerase. Each PCR tube contained a total volume 25μl, which was comprised of:
- 5μl buffer
- 0.5μl dNTP
- 0.25μl phusion polymerase
- 16.25μl H2O
- 1μl of each primer
- 1μl of template DNA
We used gel electrophoresis to check the results and isolate our fragments.
Gibson Assembly
In order to combine the fragments of the GFP containing fragment and our chosen sequence of interest (SrfA promoter) we performed Gibson Assembly.
The amplified fragments from the PCR reaction above were added into a PCR tube with a Gibson assembly master mix
| Quantity (1μl) | Reagent |
|---|---|
| 1μl | Amplified SrfA fragment |
| 1μl | Amplified Vector fragment 1 |
| 1μl | Amplified Vector fragment 2 |
| 9μl | Master Mix |
The thermocycler was set at 50 degrees C for 1 hour.
Transformation of E. coli and amplification
Following Gibson Assembly we transformed E.coli with the DNA we had combined. See the protocols page for how to do Transformation of E.coli. We transformed and plated our E. coli on a medium containing ampicillin and left them to incubate overnight. In the morning we observed 5 colonies on our plate.
To our surprise, the E. coli colonies did exhibit fluorescence when viewed under a light microscope - the B. Subtilis promoter has cross-species activity.
Digestion with Restriction Enzymes
In order to confirm correct assembly of the plasmid, we performed restriction mapping. At first, we extracted DNA from E.coli colonies that had been grown overnight in liquid medium (we used QIAGEN MiniPrep Kit technique ). The next step involved incubation with KpnI and PstI restriction enzymes, according to the following protocol.
In this experiment we performed digestion of three different samples:
- the plasmid with fusion of SrfA promoter with GFP
- the positive control plasmid with mRUBY sequence replacing GFP coding sequence
- the original pJS104 plasmid
The two restriction enzymes we chose, KpnI and PstI, were predicted to:
- give products within the 1-3kb range
- give different patterns for the three DNA samples analysed (so that we would be able to distinguish between the original and modified plasmids).
The predicted restriction maps, prepared using ApE editor, are presented below.
We separated products of digestion by gel electrophoresis (using 1% agarose gel). As we can see in the picture on the left, we did not obtain clear bands. This was most probably caused by poor quality of the gel or unfavourable concentration of the buffer. Additionally, the number of smeared bands and their distribution is not consistent with predictions from ApE, and on some lanes with uncut vectors (e.g. lane 14) we can see several bands of different sizes, although a single band was expected.
A table describing the content of each lane (numbered from the left to the right):
| Lane | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| Sample | DNA ladder | pJS104 plasmid | pJS104 plasmid | positive control | positive control | SrfA:GFP fusion A colony | SrfA:GFP fusion A colony |
| uncut | cut | uncut | cut | uncut | cut | ||
| 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 |
| SrfA:GFP fusion B colony | SrfA:GFP fusion B colony | SrfA:GFP fusion C colony | SrfA:GFP fusion C colony | SrfA:GFP fusion D colony | SrfA:GFP fusion D colony | SrfA:GFP fusion E colony | SrfA:GFP fusion E colony |
| uncut | cut | uncut | cut | uncut | cut | uncut | cut |
References
Sporulation occurs late in the life cycle of B. subtilis when the colony reaches a high population density and we hope that GFP could be visualised following overnight growth. This paper[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1178101/?page=2] details some sporulation inducing culture conditions.
 "
"